A machine learning model for cell type classification?
When analyzing single-cell transcriptomic data, scientists often perform cell type annotations by checking individual marker genes. However, marker genes are not even consistent among different literature sources. Six months ago, armed with the largest curated single-cell transcriptomic data, BioTuring single-cell team naively thought that we could solve the cell type annotation problem, simply by building a machine learning model for predicting cell type. We also thought that the machine learning model can help scientists recognize not only cell types, but also cell states, cell conditions (disease/control/etc.)
We kickstarted this project, and our initial excitement quickly turned into nightmares…
What were the reasons?
- Annotations in published studies are not consistent. Even with the same cell, different research groups can annotate it with different labels, either with a general cell type or a very specific subtype — depending on their research goals or even opinions!!
- There are many rare cell populations not having enough data points for learning. For example, the new 30 AXL+SIGLEC6+ dendritic cells identified by Villani and colleagues (Villani et. al., 2017) will be very difficult to incorporate in any machine learning models built from millions of other common cell types.
We failed in the path of building a machine learning model for predicting cell types and the enormous effort of a 4-engineer team in 6 months could be wasted!
Desperately, we thought we were probably not smart enough!
Or the nature of the problem we initially formulated is intrinsically difficult?

When we are desperately stuck, usually, there are some moments we ask ourselves: how would some of our former teachers/advisors solve the problem? A great example came back: When Mike Waterman and Pavel Pevzner faced the difficult Hamiltonian problem in genome assembly, they instead reformulated the problem to an Eulerian path problem, which can be solved efficiently (https://www.pnas.org/content/98/17/9748.short).
Reformulating the problem as a cell search problem.
Given the largest indexed single-cell data, we imagine, when a scientist selects a group of cells, a cell search engine can help find all cells in all published studies that have “similar” expression signatures, together with their cell type labels. An important difference of the prediction model and the search problem is that the former takes subjective human annotations as input to the model, while the latter does not. The search engine allows human annotations to be verified by human! This helps bypass the challenges.
After getting results from a search operation, scientists can download the matched cells, and see all other labelings of these cells. These may include age, disease, tumor/normal conditions. For instance, would it be more interesting to see that this group of microglia cells only appears in Parkinson patients rather than normal?
An important challenge in this cell search engine is that it has to bypass technical variations (cells with different biological conditions but were sequenced under similar sequencing technologies) to return only the cells that match biological conditions. We successfully solved this problem (details will be described in a future manuscript).
BioTuring Cell Search: a novel search engine for single-cell RNA-seq data
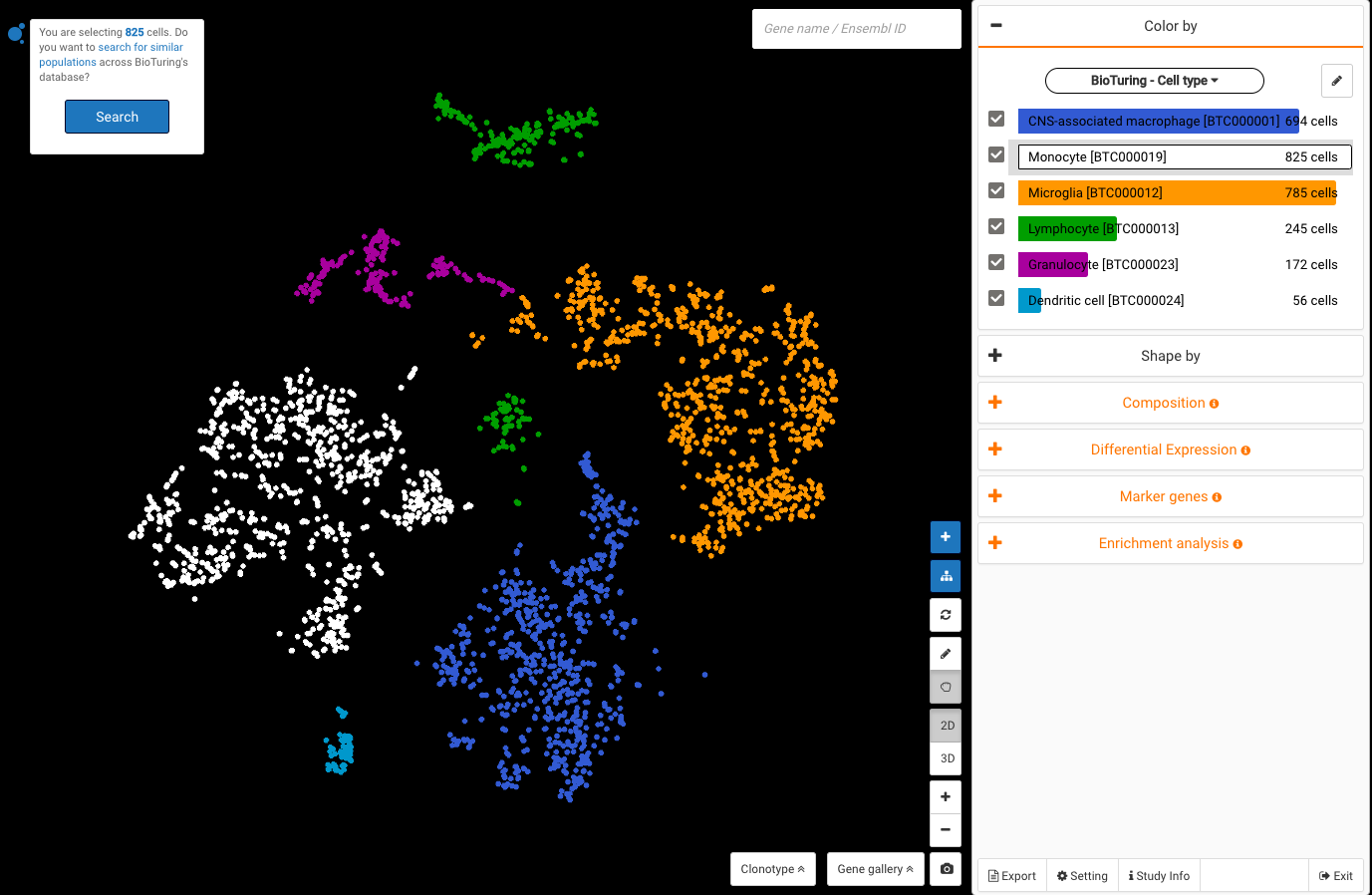
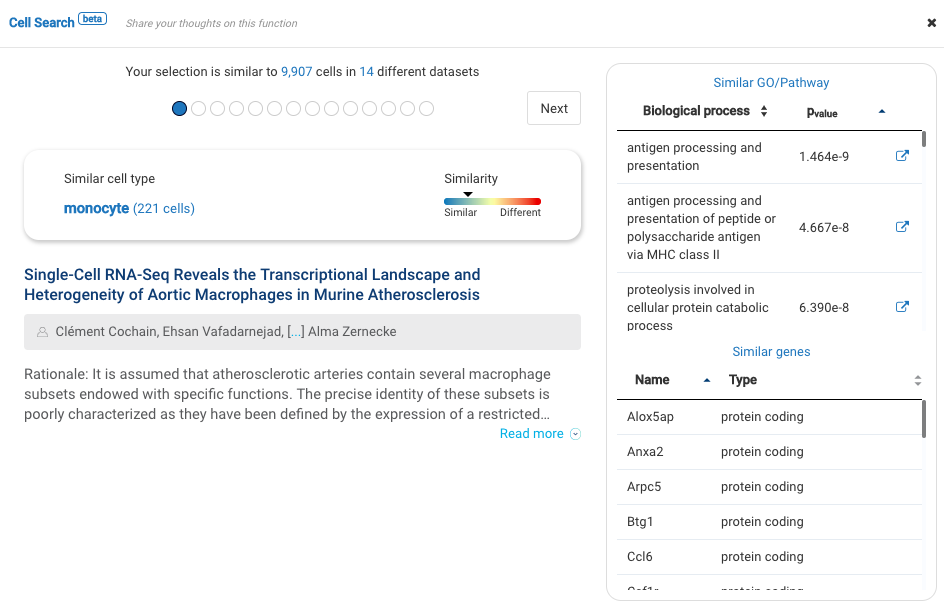
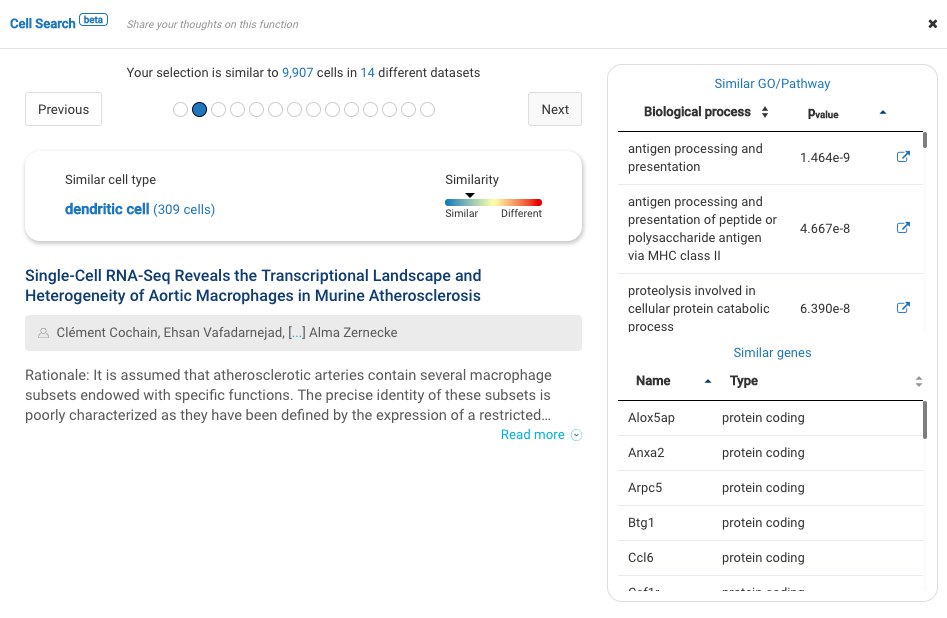
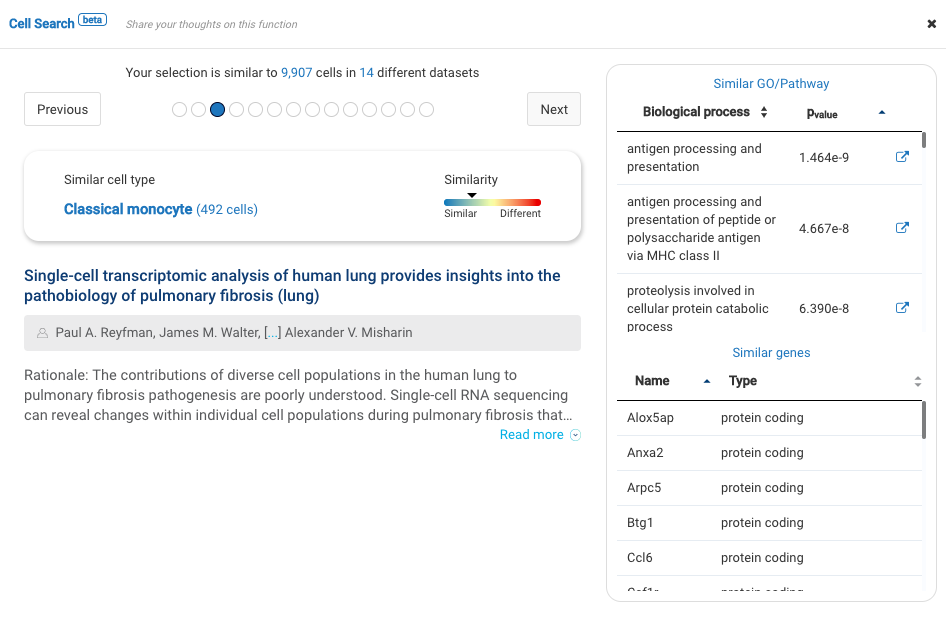
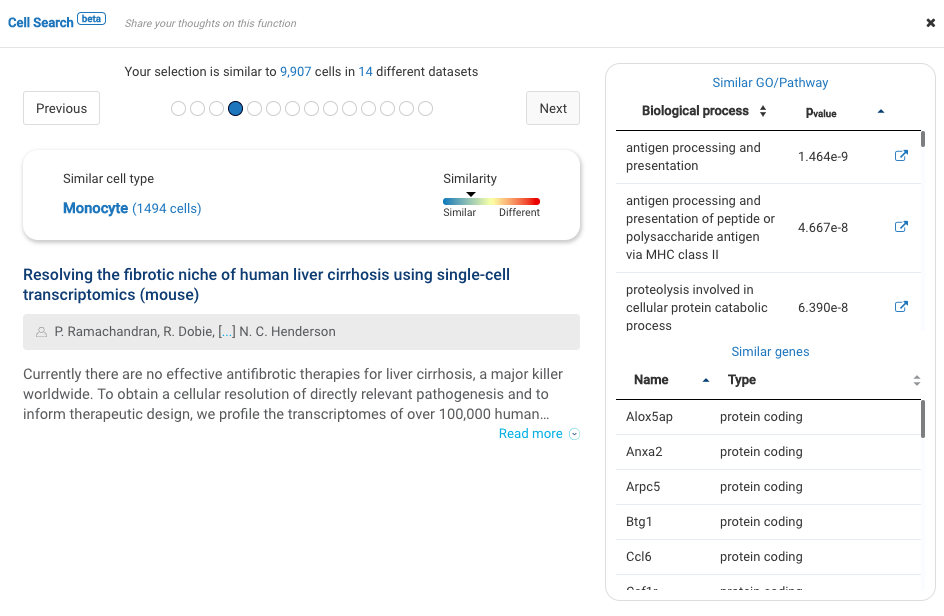
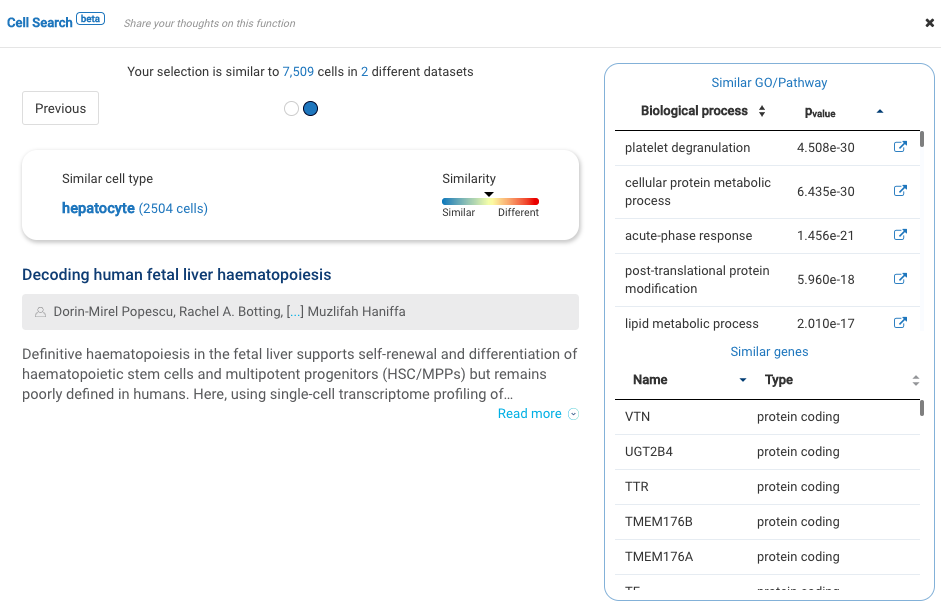
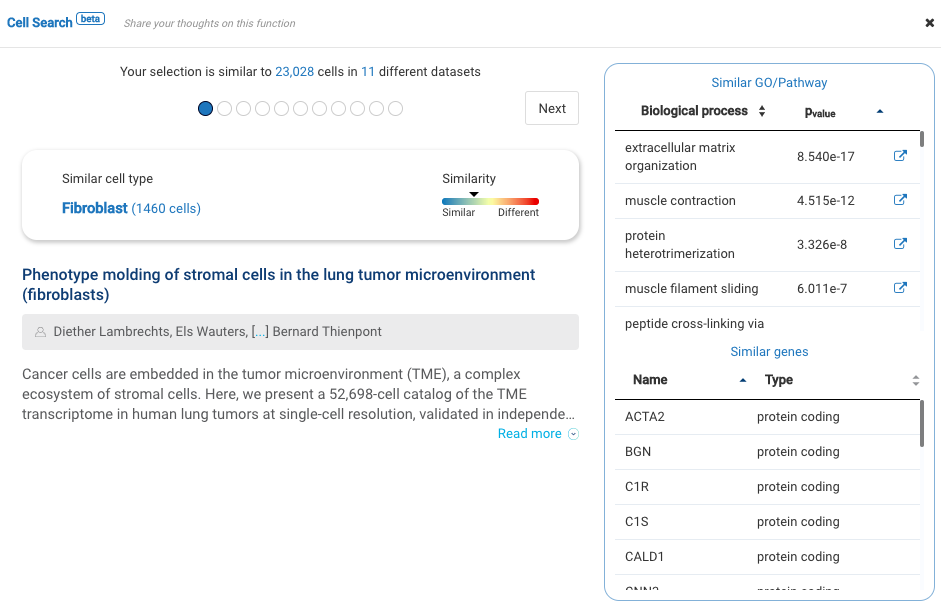
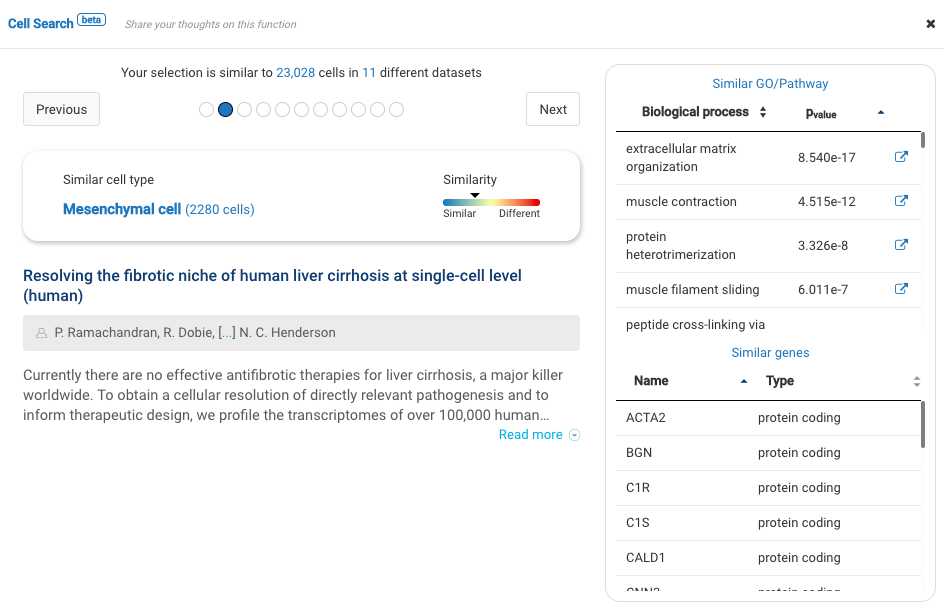
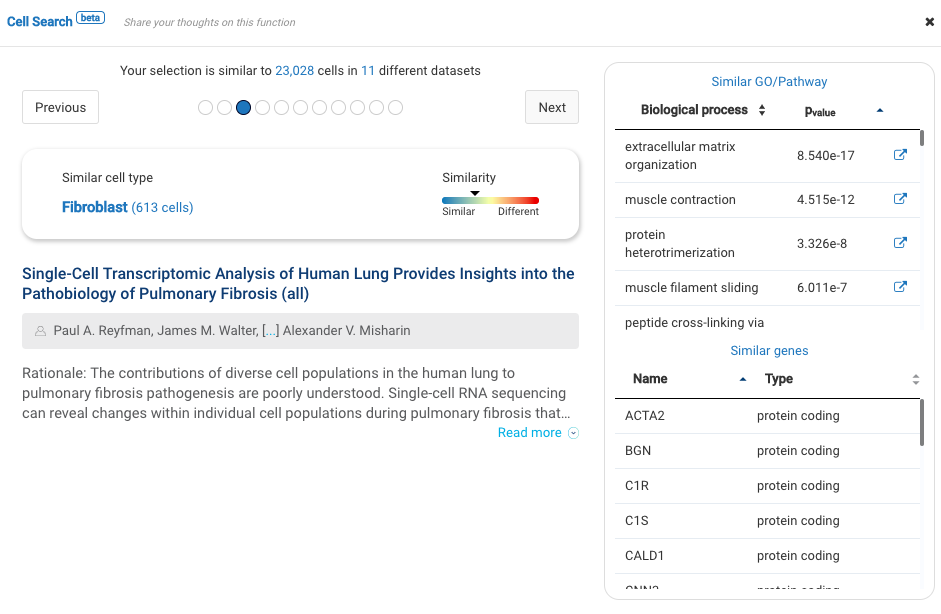
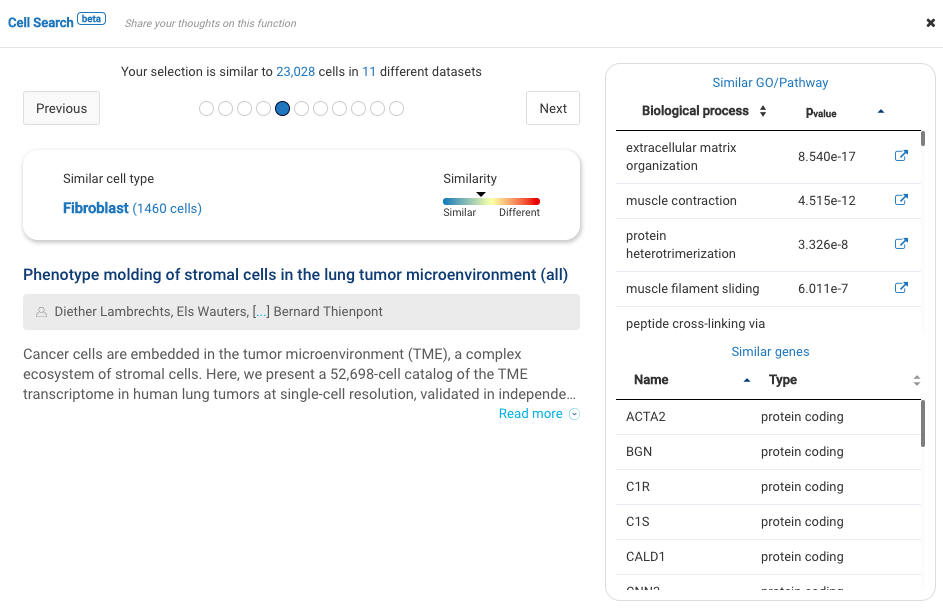
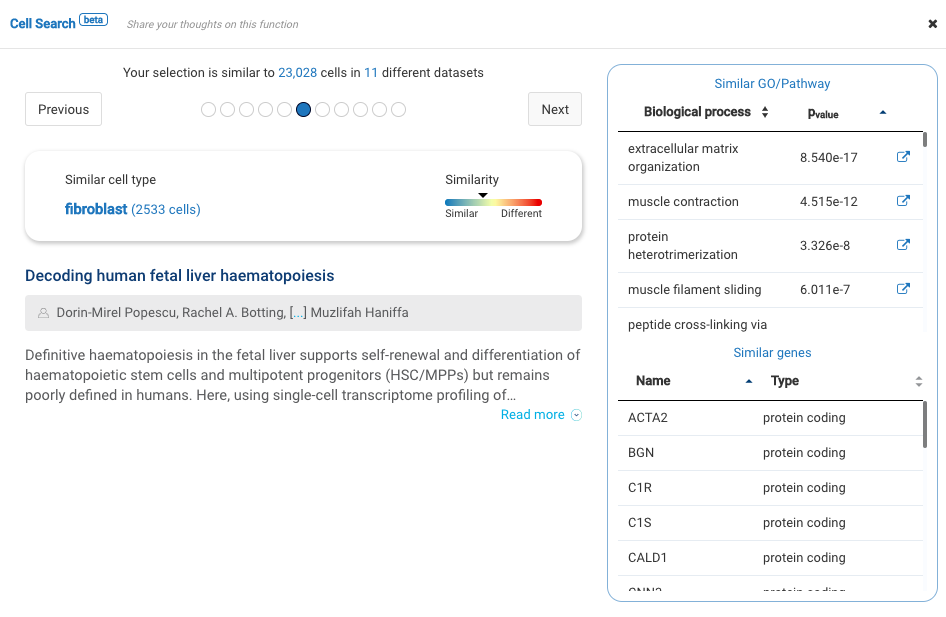
Our team built and launched BioTuring Cell Search, a search engine that enables quick and accurate searching of similar cells across our single-cell database of 5 million cells, curated from more than 125 publications. Upon the selection of a group of cells, scientists will get:
- A list of published studies with matched populations and their cell type labelings
- Similarity scores between the gene expression profiles of matched populations and the selected cells
- Similarly expressed genes and enrichment processes shared across all matched populations
Based on the search results, scientists can download the data sets with matched cells, study their states, conditions, compositions, and other annotations, and finally come back to annotate their cells at their own discretion.
Example: Using BioTuring Cell Search to verify cell type identification results
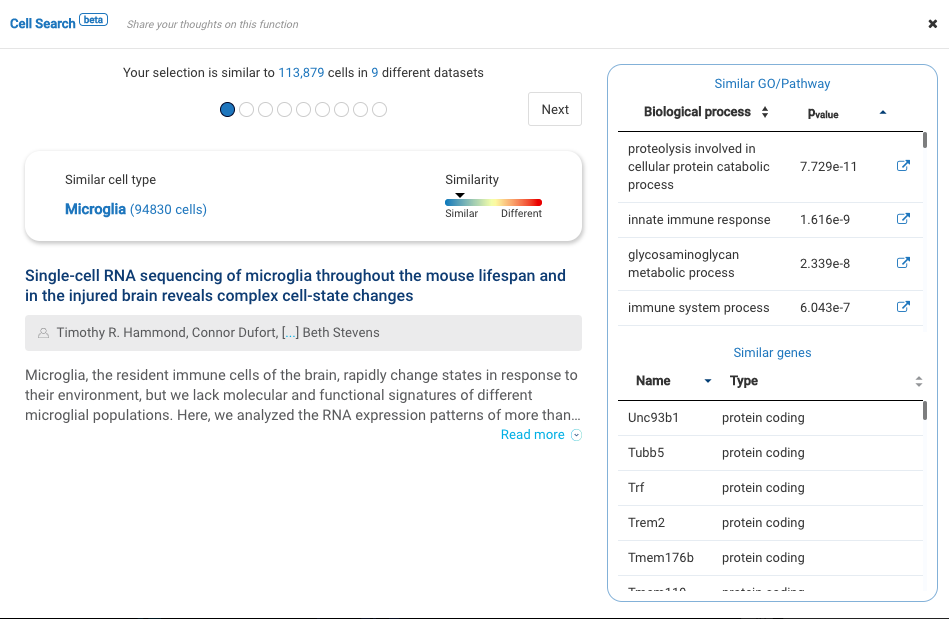
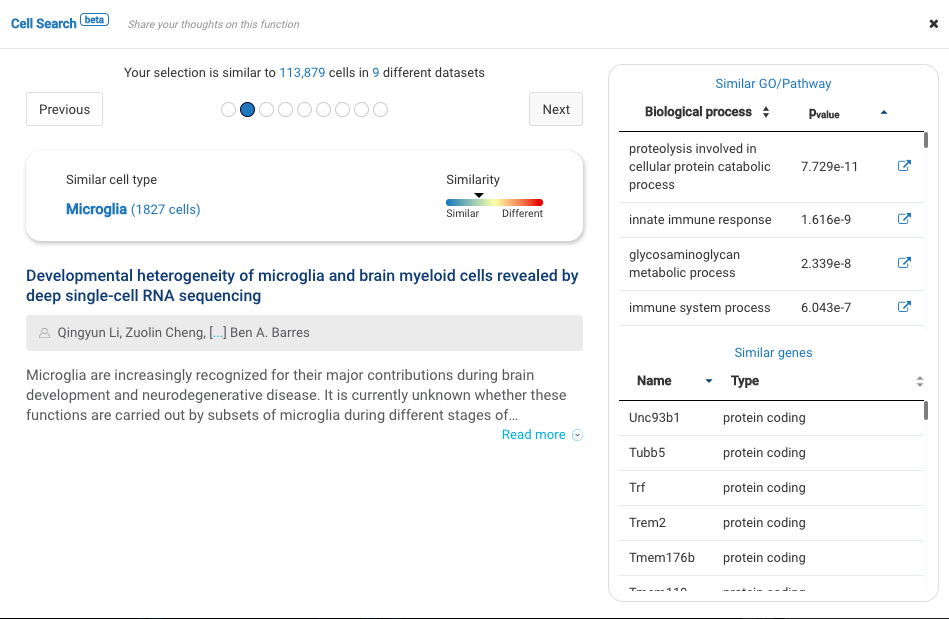
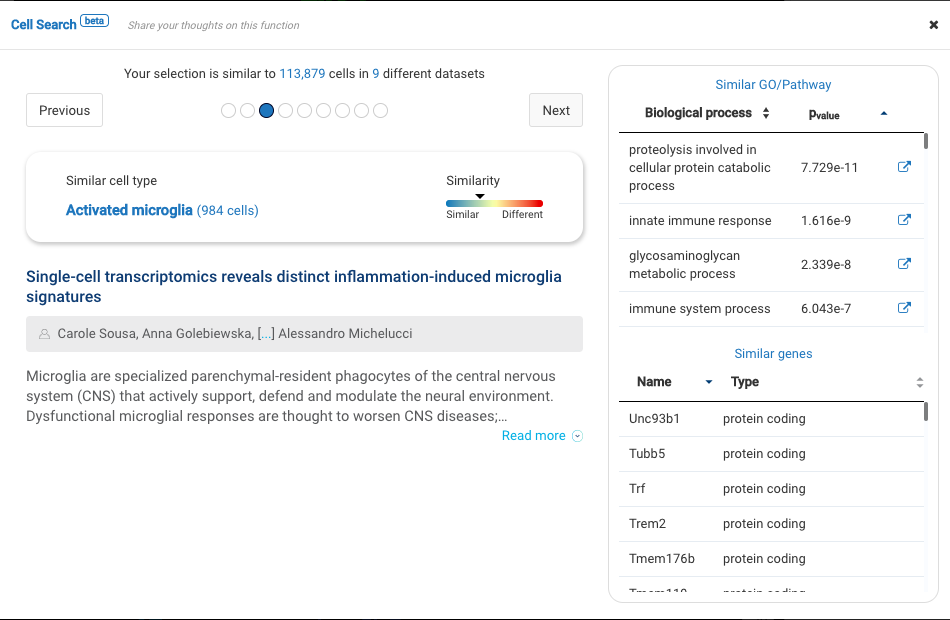
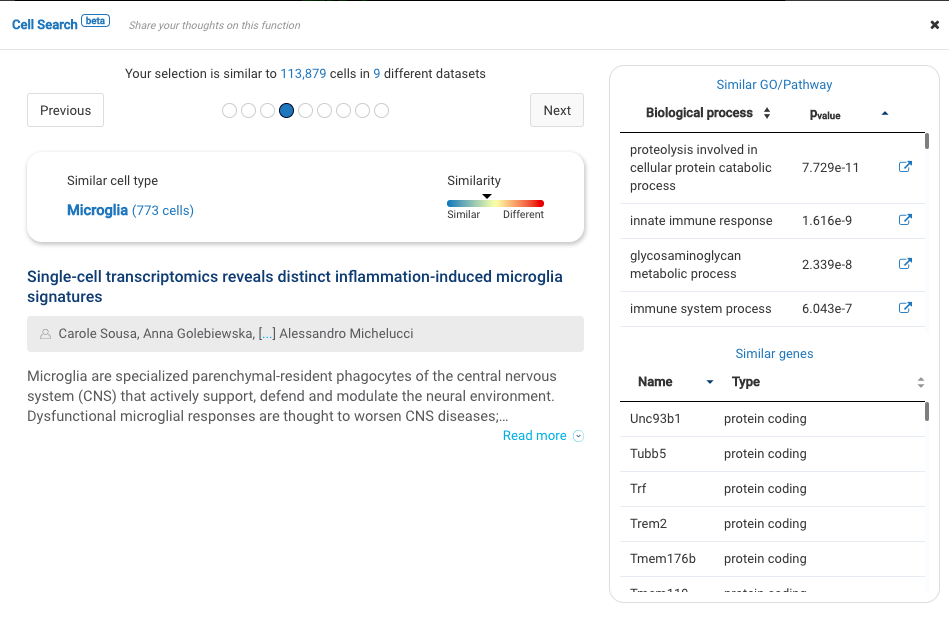
Dataset 1: Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation (Jordao et al., 2019)
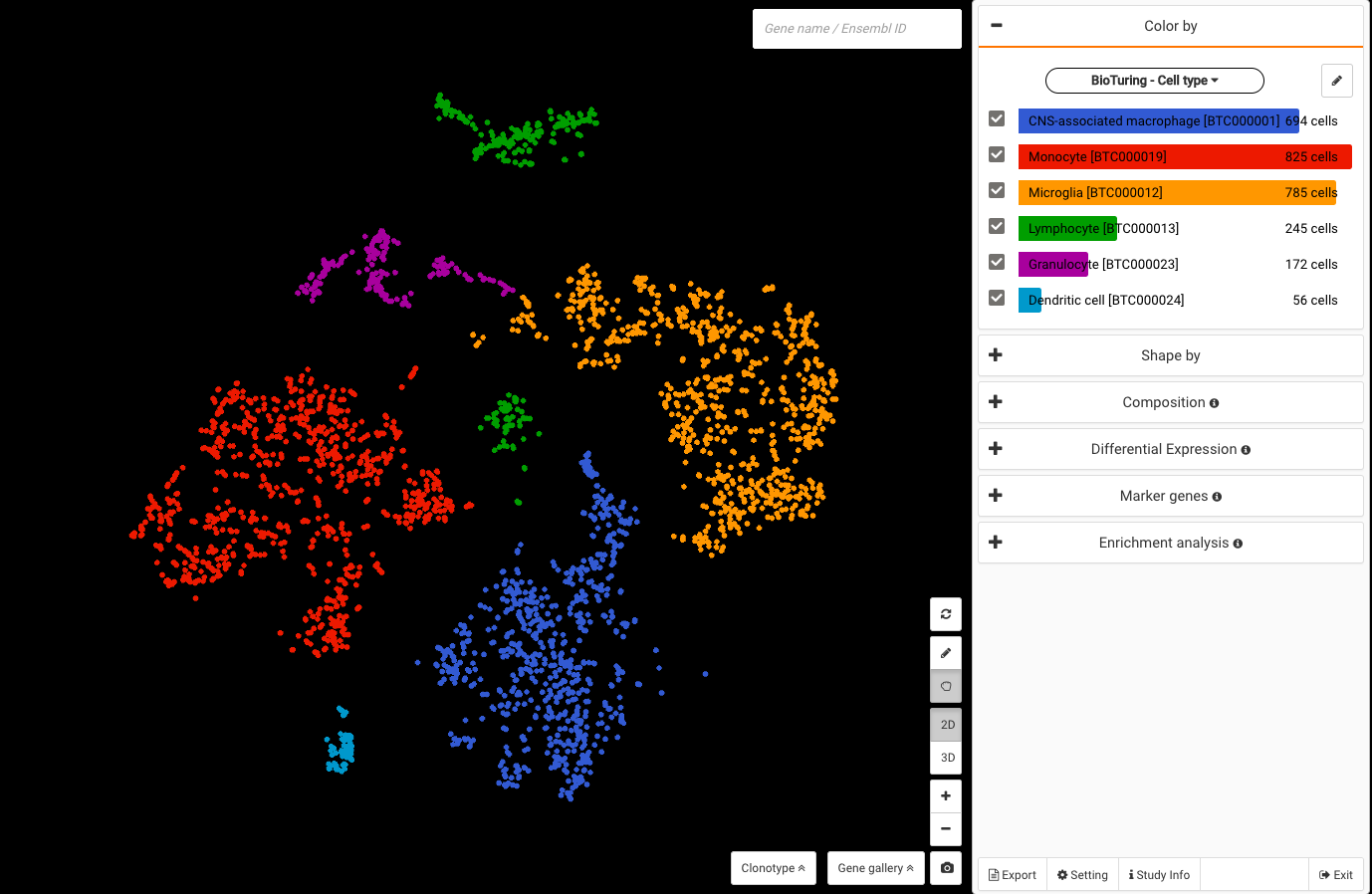
Profiling more than 3000 myeloid cells in the central nervous system (CNS) in multiple sclerosis mouse models, the study provided an atlas of myeloid cells and their dynamics across various stages of neuroinflammation. Major cell types were identified, including microglia, lymphocytes, CNS associated macrophages, dendritic cells, granulocytes, and monocyte-derived cells.
BioTuring Cell Search results on each cluster of the data confirm the cell types recognized by the study.
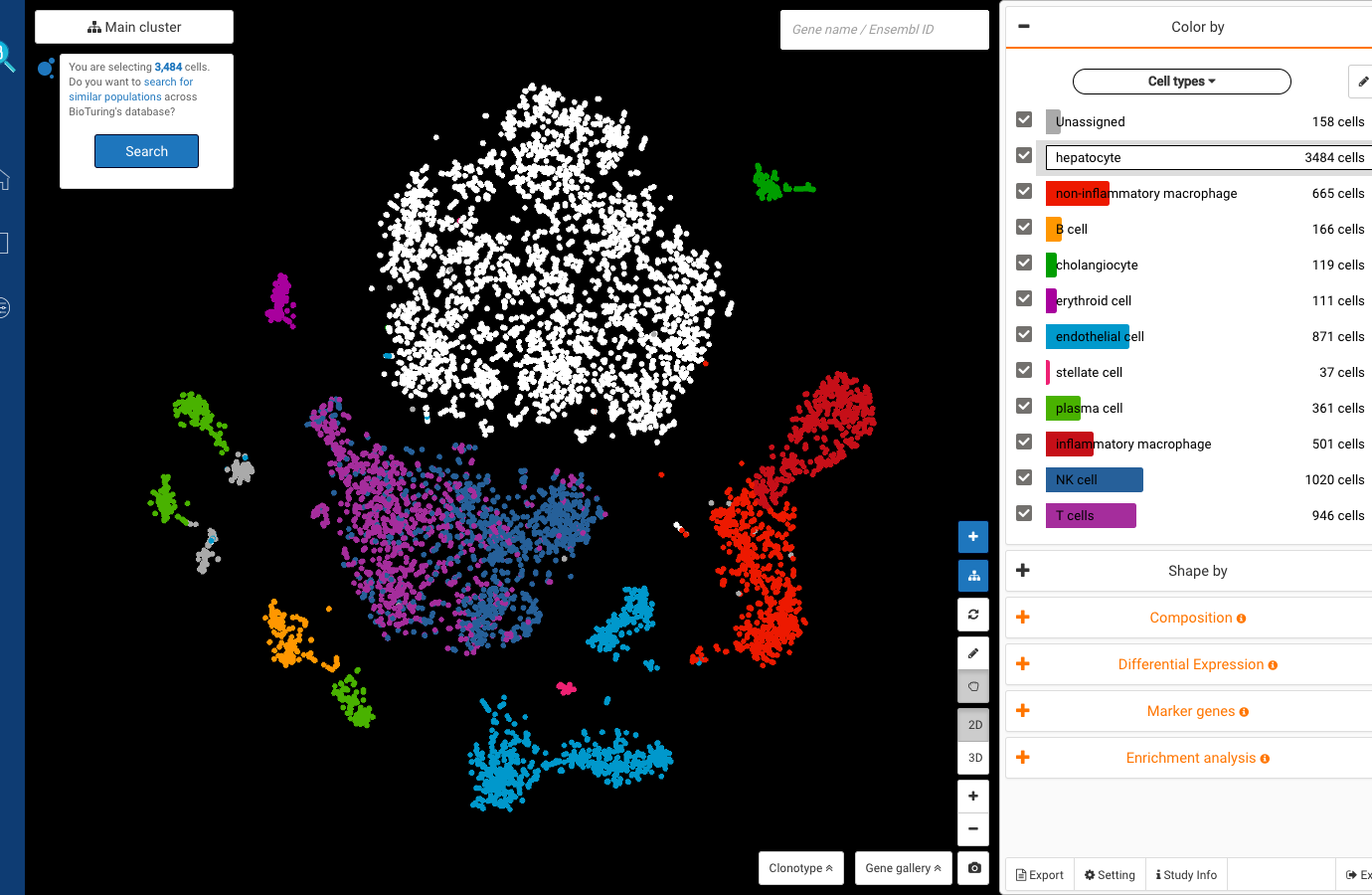
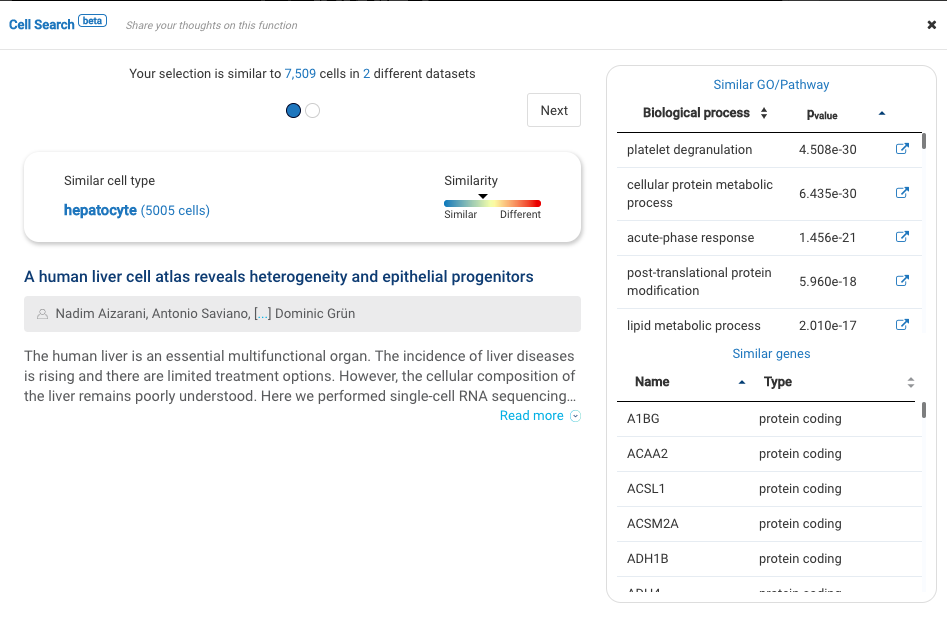
Dataset 2: Single-cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations (MacParland et al., 2018)
Published in 2018, the work by MacParland and colleagues is one of the very first human liver cell atlases, unveiling new understandings into the liver cellular heterogeneity.
With BioTuring Cell Search, we sought to verify the cell types in the dataset. Most labels match previous publications, including B cells, hepatocytes, endothelial cells, plasma cells, and natural killer cells.
Other cell types like cholangiocytes and macrophages match various populations in the database, yet with different cell type labels. Stellate cells, meanwhile, exhibit some similarity level with fibroblasts. The population highly expresses genes encoding for collagen production (COL1A2 and COL3A1).
BioTuring Cell Search now can be used with BioTuring Browser, an intuitive platform for exploring single-cell transcriptomic data. The platform is available for download at https://bioturing.com. It can also be called via API.
—
Reference:
Jordão, Marta Joana Costa, et al. "Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation." Science 363.6425 (2019): eaat7554.
MacParland, Sonya A., et al. "Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations." Nature communications 9.1 (2018): 4383.
Pevzner, Pavel A., Haixu Tang, and Michael S. Waterman. "An Eulerian path approach to DNA fragment assembly." Proceedings of the national academy of sciences 98.17 (2001): 9748-9753.
Villani, Alexandra-Chloé, et al. "Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors." Science 356.6335 (2017): eaah4573.