BIRC5, known as Survivin, is one member of the Inhibitor of apoptosis protein family and plays an important role in preventing cell death (Jaiswal et al., 2015) and poor survival in cancers (Supplementary). For this reason, BIRC5 can be hypothesized to be associated with tumor resistance to cancer treatments.
With the largest single-cell database in BioTuring Browser, we want to verify this hypothesis.
1. BIRC5 is up-regulated in the tumors that resist to Immune Checkpoint Inhibitors (ICIs) in Melanoma
We first checked BIRC5 expression in the single-cell data set by Jerby-Arnon et al. (2018). This data set contains melanoma cells from 2 groups of patients: resistant to ICIs (combining multiple therapies like CTLA-4, PD-1, PD-L1 and KIR blockade) and untreated patients (See Table 1).
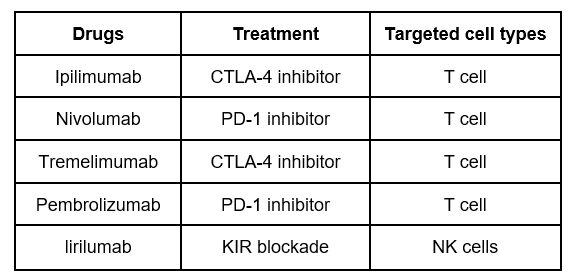
Table 1. Multiple immune checkpoint inhibitors (ICIs) and their targeted cells in the single-cell data set by Jerby-Arnon et al. (2018)
We compared BIRC5 expression in malignant cells of ICI resistance vs. untreated groups. Interestingly, BIRC5 was significantly up-regulated in ICI resistance (Fig.1).
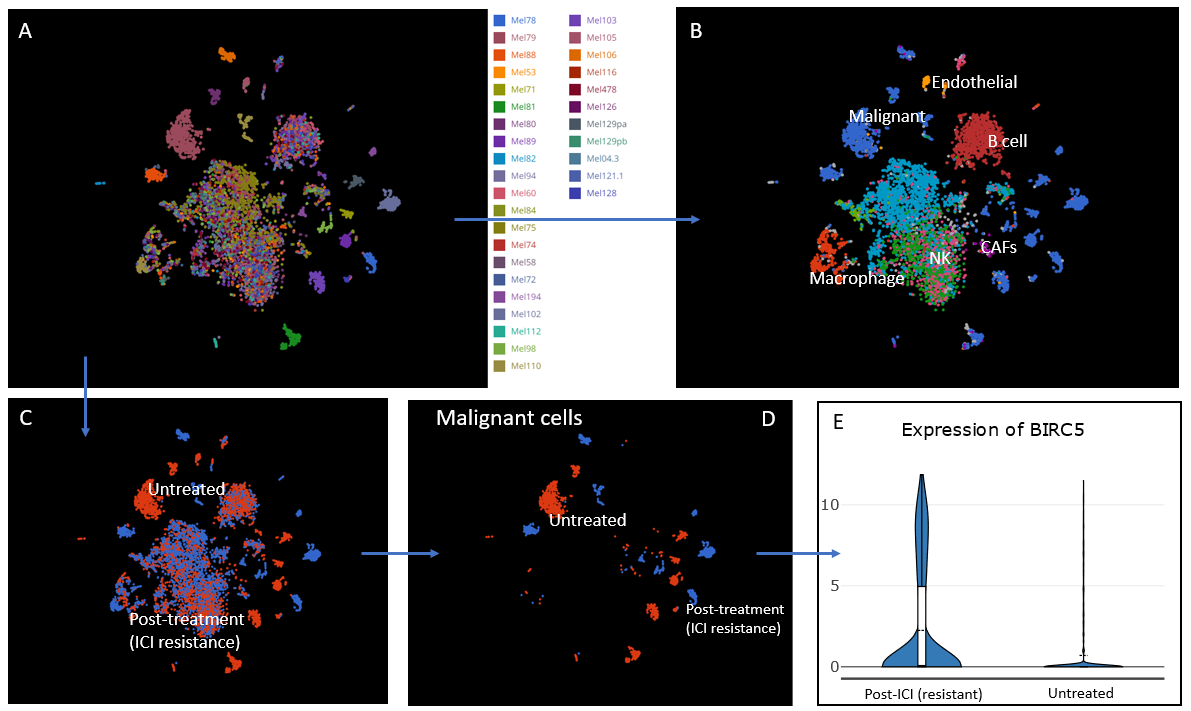
Figure 1. (A) All patients in single-cell data set from Jerby-Arnon et al. (2018) in BBrowser. (B) Cell types from all patients. (C) Two groups Untreated and Post-treatment (ICI resistance). (D) Malignant cells with two groups Untreated and ICI resistance. (E) In malignant cells of melanoma, BIRC5 is highly expressed in ICI resistance in comparison with Untreated group.
2. BIRC5 expression increases with the levels of resistance in CDK4/6 inhibition therapy in Melanoma
In addition to ICIs, Jerby-Arnon et al. also profiled the melanoma cell lines in response to CDK4/6 inhibition therapy (Abemaciclib), which inhibits cell cycle at G1/S phase.
In malignant cells, we found BIRC5 expression significantly increased with a higher level of resistance to Abemacilicb therapy. This did not happen to the control groups (DMSO) (Fig.2).
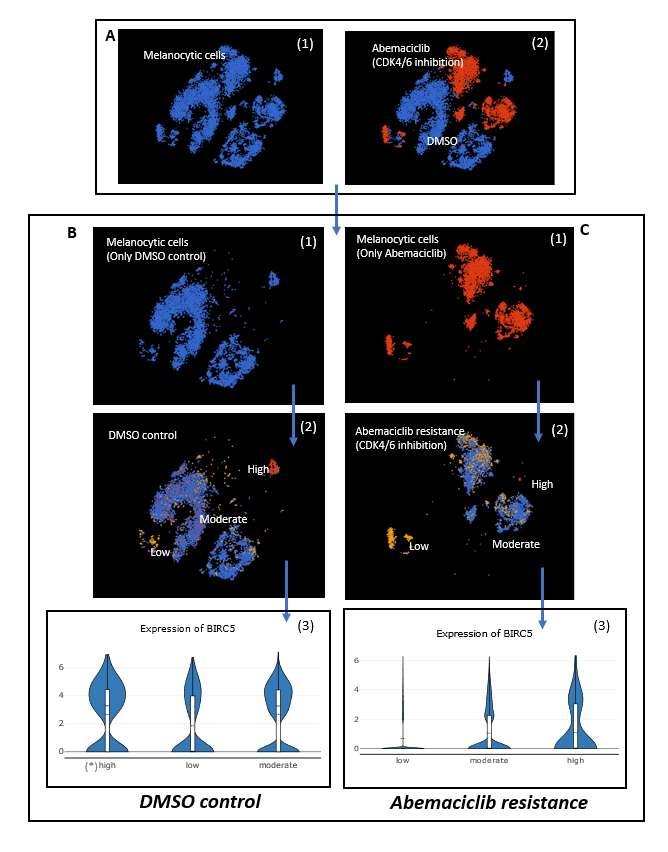
Figure 2. (A) Melanocytic cells with CDK4/6 inhibition (Abemaciclib) and DMSO control. (*) Jerby-Arnon et al. defined the low to high resistant groups if they underexpressed one of the top specific genes: B2M, CD58, HLA-A, MLANA, SOX10, SRP54, TAP2 and TAPBP (Lowest level: 1%). (B) In DMSO control of melanocytic cells, the level of BIRC5 expression did not change from low to high resistance. (C) In Abemaciclib resistance, BIRC5 expression significantly increased from low to high resistance with the therapy.
3. BIRC5 is up-regulated in chemo-resistance in Oral cancer
We further explored the data set of Sharma et al. (2018) to see if BIRC5 correlates with chemotherapy (Cisplatin) resistance. In chemo-resistance tumor cells, BIRC5 was up-regulated, compared with drug-sensitive and drug-holiday samples (Fig.3).
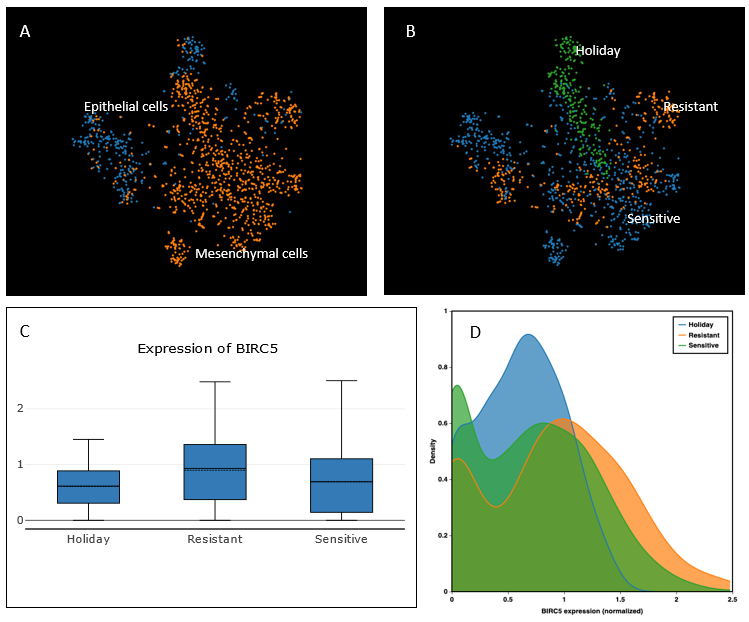
Figure 3. In oral cancer, Sharma et al. (2018) collected (A) Epithelial cells and Mesenchymal cells. (B) The clusters with Chemotherapy (Cisplatin) resistance: Sensitive, Holiday and Resistant. (C) Comparison among BIRC5 expressions in drug-responses. (D) The density of BIRC5 expression with three drug-response groups.
So what’s the “verdict”? After a quick analysis on BioTuring database, BIRC5 seems to be linked with tumor resistance at single-cell resolution. A more thorough analysis will be required for a stronger conclusion. We suggest future works into BIRC5 to deepen our understanding of its dynamics in tumor resistance and whether blocking BIRC5 proves to be an effective therapeutic approach for treating cancers.
Supplementary
In cell cycle, Survivin is also an essential mitotic protein to control cell division and apoptosis (Fengzhi et al., 2006), especially expressed in G2/M (Chandele et al., 2004).
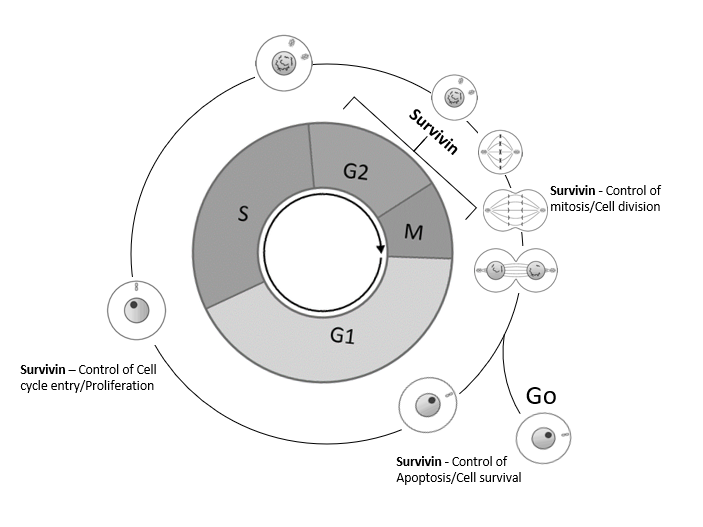
Fig S1. BIRC5 (Survivin) controls mitosis and cell division (G2/M → G1), apoptosis/cell survival (Go/G1 or G1 to cell death) and cell cycle entry/proliferation (Go/G1 → S/G2) (Fengzhi et al., 2006), especially expressed in G2/M (Chandele et al., 2004).
As shown in previous studies have shown BIRC5 as a prognostic marker for poor survival in many types of cancer (Chade et al., 2018, Cheng et al., 2016, Contis et al., 2018, Jaiswal et al., 2015), inducing resistance to apoptosis and higher chances for relapse (Rodel et al., 2012, Islam et al., 2000).
This article is for research purpose only.
References
- Jaiswal, P. K., Goel, A., & Mittal, R. D. (2015). Survivin: A molecular biomarker in cancer. The Indian journal of medical research, 141(4), 389–397. https://doi.org/10.4103/0971-5916.159250
- Fengzhi Li and Michael G. Brattain (2006). Review: Role of the Survivin Gene in Pathophysiology. American Journal of Pathology, Vol. 169, No. 1, Copyright © American Society for Investigative Pathology. DOI: 10.2353/ajpath.2006.060121
- Chandele, A., Prasad, V., Jagtap, J. C., Shukla, R., & Shastry, P. R. (2004). Upregulation of Survivin in G2/M cells and inhibition of caspase 9 activity enhances resistance in staurosporine-induced apoptosis. Neoplasia (New York, N.Y.), 6(1), 29–40. https://doi.org/10.1016/s1476-5586(04)80051-4
- Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y, Nakagawara A (2000) High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene; 19(5):617-23. DOI: 10.1038/sj.onc.1203358
- Rodel F, Sprenger T, Kaina B, Liersch T, Rödel C, Fulda S, Hehlgans S (2012) Survivin as a prognostic/predictive marker and molecular target in cancer therapy. Curr Med Chem;19(22):3679-88. DOI: 10.2174/092986712801661040
- Chade MC, Piato S, Galvão MAL, Aldrighi JM, Negrini R, Mateus EF, Medeiros EM (2018), Evaluation of survivin immunoexpression in the differentiation of high- and low-grade breast ductal carcinoma in situ, Einstein (Sao Paulo); 16(1):eAO4065. DOI: 10.1590/s1679-45082018ao4065
- Contis J, Lykoudis PM, Goula K, Karandrea D, Kondi-Pafiti A (2018), Survivin expression as an independent predictor of overall survival in pancreatic adenocarcinoma, J Cancer Res Ther;14 (Supplement): S719-S723. DOI: 10.4103/0973-1482.187346
- Jerby-Arnon, L., Shah, P., Cuoco, M. S., Rodman, C., Su, M. J., Melms, J. C., Leeson, R., Kanodia, A., Mei, S., Lin, J. R., Wang, S., Rabasha, B., Liu, D., Zhang, G., Margolais, C., Ashenberg, O., Ott, P. A., Buchbinder, E. I., Haq, R., Hodi, F. S., … Regev, A. (2018). A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell, 175(4), 984–997.e24. https://doi.org/10.1016/j.cell.2018.09.006
- Sharma A, Cao EY, Kumar V, et al. Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy (2018). Nat Commun ;9(1):4931. doi:10.1038/s41467-018-07261-3