Take a non-small cell lung tumor. What do we see?
To answer this challenging question, a lot of single-cell omics experiments have been conducted, yielding significant insights into the heterogeneity of non-small cell lung cancer (NSCLC) microenvironment. While each successfully characterizes a facet of this ecosystem, until now no work has been done to compile these works and build a comprehensive atlas characterizing all cell types and subtypes found in NSCLC.
Putting pieces of the puzzle together
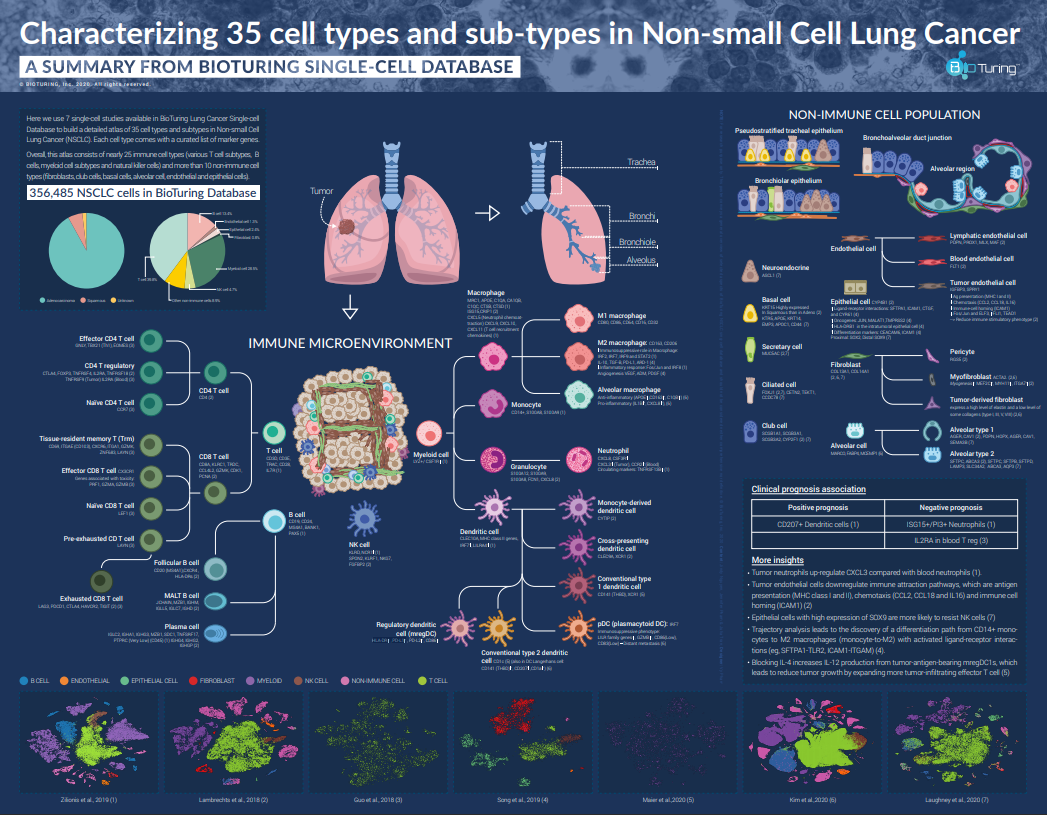
We sort out 356,485 cells from 7 studies on non-small cell lung cancer (NSCLC) (Zilionis et al., 2019; Lambrechts et al., 2018; Guo et al., 2018; Song et al., 2019; Maier et al.,2020; Kim et al.,2020) in BioTuring Single-cell Database and compile a complete atlas of NSCLC cell types and subtypes found in all those studies.
Get a full .pdf version of the poster
The final atlas consists of 25 immune and 10 non-immune cell types and subtypes (e.g. T cells, B cells, myeloid cells, natural killer cells, fibroblasts, club cells, basal cells, alveolar cells, endothelial, epithelial cells,…). Each cell type comes with a curated list of marker genes, which we have cross-validated across all lung single-cell datasets in the BioTuring database.
All immune cells found in BioTuring NSCLC Single-cell Database
Please view our poster for a full list of cell types, subtypes, and markers
|
Major cell types |
Cell types – Subtypes |
References |

T cell (Markers: CD3D,CD3E, TRAC, CD28 IL7R) |
CD4 T cell (Markers: CD4)
|
(1,2,3) |
CD8 T cell: (Markers:CD8A, GZMK…)
|
(2,3) |
|
 B cell B cell
(Markers: CD19, CD24, MS4A1, BANK1,PAX5) |
|
(1,2) |
 Myeloid cell Myeloid cell
(Markers: LYZ+, CSF1R+) |
Macrophage (Markers:MRC1, APOE…)
|
(1,2,4,5,6) |
Granulocyte (Markers:S100A12, S100A9…)
|
(1,2) |
|
| Monocyte (Markers: CD14+, S100A8…) |
(1) |
|
Dendritic cell (Markers:CLEC10A, MHC class II genes…)
|
(1,2,5,6) |
|

NK cell (Markers: KLRD, NCR1+, SPON2, KLRF1, NKG7, FGFBP2) |
(2) |
|
“+” indicates high expression levels.
Non-immune cells found in BioTuring NSCLC Single-cell Database
Please view our poster for a full list of cell types, subtypes, and markers
|
Cell types |
Markers |
References |
| IGFBP3, SPRY1 | (2) | |

Epithelial cell |
CYP4B1, Proximal: SOX2, Distal SOX9 | (2,7) |
Fibroblast |
COL13A1, COL14A1 | (2,6) |

Alveolar cell (Alveolar type 1, Alveolar type 2) |
MARCO, FABP4, MCEMP1 | (6) |

Club cell |
SCGB1A1, SCGB3A1, SCGB3A2, CYP2F1 | (2,7) |
What are other insights?
From 7 studies, we also summarize some interesting insights in terms of clinical prognosis and more. See below:
- CD207+ dendritic cells are associated with positive prognosis (1), while ISG15+/PI3+ neutrophils and IL2RA+ blood T regs are associated with poor prognosis (1,3)
- Tumor neutrophils up-regulate CXCL3 compared with blood neutrophils (1).
- Tumor endothelial cells downregulate immune attraction pathways, which are antigen presentation (MHC class I and II), chemotaxis (CCL2, CCL18 and IL16) and immune cell homing (ICAM1) (2)
- Epithelial cells with high expression of SOX9 are more likely to resist NK cells (7)
- Trajectory analysis leads to the discovery of a differentiation path from CD14+ monocytes to M2 macrophages (monocyte-to-M2) with activated ligand-receptor interactions (eg, SFTPA1-TLR2, ICAM1-ITGAM) (4).
- Blocking IL-4 increases IL-12 production from tumor-antigen-bearing mregDC1s, which leads to reduced tumor growth by expanding more tumor-infiltrating effector T cells (5)
Above is our sketch of the immune and non-immune cells found in NSCLC, and their prognostic significance, compiled from multiple single-cell RNA-seq datasets. We suggest further work to characterize multiple facets of this environment to provide more detailed background information for NSCLC clinical directions.
This article is for research purposes only.
References
1. Zilionis et al., Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity, 2019. Volume 50, Issue 5, https://doi.org/10.1016/j.immuni.2019.03.009.
2. Lambrechts et al., Phenotype molding of stromal cells in the lung tumor microenvironment. Nature Medicine 2018. 24, pages 1277–1289 https://doi.org/10.1038/s41591-018-0096-5.
3. Guo et al., Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nature Medicine, 2018. 24, 978–985 https://doi.org/10.1038/s41591-018-0045-3.
4. Song et al., Dissecting intratumoral myeloid cell plasticity by single-cell RNA-seq. Cancer Med, 2019. 8(6):3072-3085. https://doi.org10.1002/cam4.2113.
5. Maier et al., A conserved dendritic-cell regulatory program limits antitumor immunity. Nature, 2020. 580, pages257–262 https://doi.org/10.1038/s41586-020-2134-y.
6. Kim et al., Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun, 2020. 8;11(1):2285. https://doi.org/10.1038/s41467-020-16164-1
7. Laughney et al., Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med 2020. 26, 259–269 https://doi.org/10.1038/s41591-019-0750-6.