1. An atlas of acute myeloid leukemia at single-cell resolution
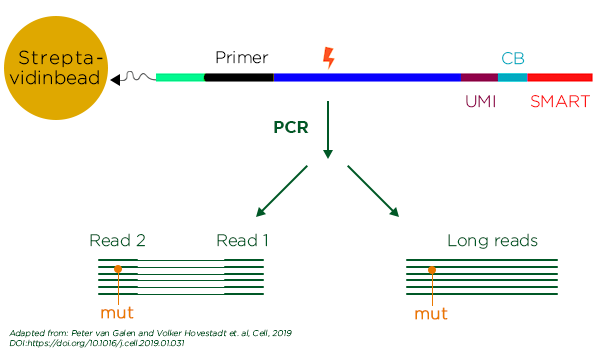
Adapted from Galan et. al., 2019
Acute myeloid leukemia (AML) is a cancer characterized by the accumulation of white blood cells in the bone marrow and blood. The disease has been notoriously challenging to study because of its extreme intratumoral heterogeneity. The complexity of AML has become possible to be tackled with the advent of high-throughput high-dimensional single-cell sequencing technologies. In a study published on Cell last month, Galen and Hovestadt et al. combined these cutting-edge technologies to gain remarkable insights into AML.
Single-cell RNA sequencing (scRNA-seq) technology can capture entire transcriptomic profile of a single cell. The authors employed high-throughput scRNA-seq to characterize the whole transcriptome of nearly forty thousand bone marrow derived cells from healthy donors. Basing on transcriptomic profile of these normal BM cells, they attempted to distinguish malignant cells and normal cells from AML patients. Unfortunately, the gene expression profile of malignant cells resembled to that of normal ones, making the attempt of separating the two groups very challenging. However, AML cells harbor a set of mutations, which can be used as genetic markers. Therefore, to address the aforementioned challenge, the author combined scDNA seq (genotyping) that can detect genetic mutations and scRNA-seq (phenotyping). basing on unique molecule barcodes. The integration of genotype and phenotype data provided a clear definition of malignant and normal cells. To further improve the applicability of their single-cell genotyping protocol, the authors employed a third-generation DNA sequencing technique called nanopore sequencing. Nanopore sequencing platform is capable of sequencing long reads, allows enhanced detection of long insertions, deletions and fusion breakpoints. With the help of machine learning (random forest algorithm) on integrated transcriptional and genetic data, van Galen and Hovestadt et al. generated an atlas of different AML cell types and normal blood cell types. Remarkably, atlas allowed the classification of sub clonal populations within the tumor cells including the primitive, differentiated and in the process of differentiating. They found 6 malignant cell types of myeloid lineage. They also found cell type composition varies among patient and strongly correlated to the tumor genotype. One differentiated AML sub-population expressing markers of CD14+ monocytes was shown to be highly aggressive and capable of suppressing T cell activation. This aggressive subclone was correlated with poor patient prognosis. In contrast, the primitive AML sub-population resembled hematopoietic stem cells/progenitor cells and was less aggressive. These findings will assist the development of novel strategies for immunotherapy and personalized treatment for AML.
2. Exploring hypothalamic circuits, one neuron at time
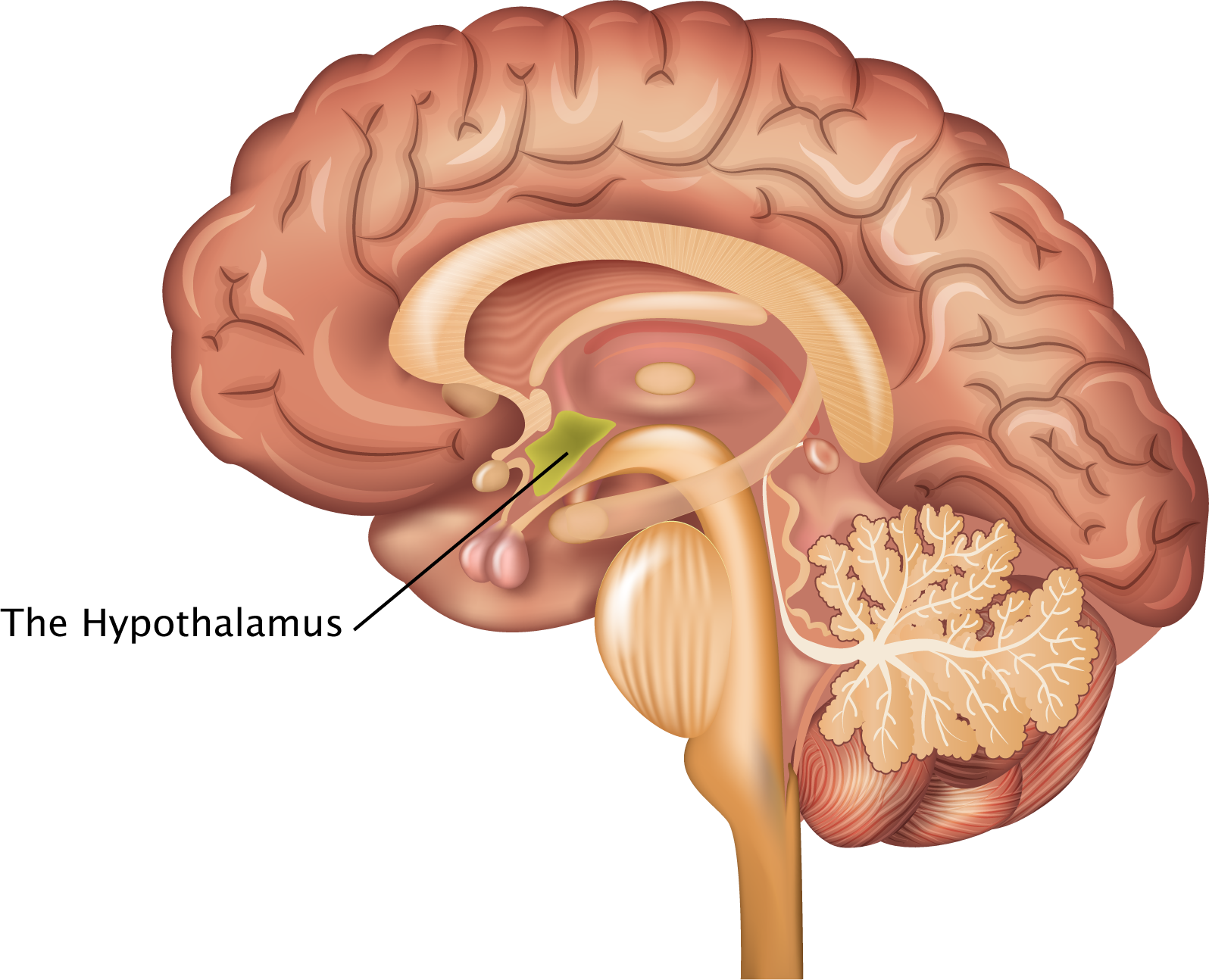
Source: thescienceofpsychotherapy.com
Despite being small part of the brain, the lateral hypothalamic area (LHA) is responsible for the regulation of metabolism, thermoregulation, wake-sleep cycles, stress responses and motivated behaviors. Yet, the comprehensive detail of neuron diversity in LHA remains undefined. In the March issue of Nature Neuroscience, Mickelsen et al. took a comprehensive census of cell types in mouse LHA using droplet-based single-cell RNA sequencing (scRNA-seq). Based on fast neurotransmitter phenotype and expression of neuropeptides, transcription factors and synaptic proteins, the author identified 15 distinct populations of glutamatergic (excitatory) neurons and 15 GABAergic (inhibitory) neurons, including known and novel cell types. The diversity helps explain various behavioral effects elicited by non-selective activation of LHA in animals, including reward-related, feeding, predatory, and other behaviors. Subsequent characterization (including anatomical distribution, electrical properties, and projection to the brain) of a novel neuron population expressing somatostatin revealed its role in locomotor behavior. These results provided foundation for future mechanistic studies of the relationship somatostatin positive neuron and activated behaviors.
3. Palantir: modeling trajectories by characterizing cell fate probabilities
Developed by Setty and colleagues at Dana Pe’er Lab, Palantir models cell trajectories by treating cell fate differentiation as a Markov process and leverages entropy to measure cell plasticity along the trajectory.
Specifically, the algorithm uses multiple diffusion components to compute the pseudo-time ordering of cells, which assigns each cell a relative distance from an initial cell, without considering its lineage or terminal fates. It then uses the nearest neighbor graph and pseudo-time to construct a Markov chain that models differentiation as a stochastic process. While pseudo-time helps with directionality and orients edges on the neighbor graph, each edge will be assigned a transition probability of reaching a neighboring cell in one step. Meanwhile, the probability of reaching a more distant cell is computed over multiple steps. If many paths connect them, the probability will be high, meaning a high density of observed intermediary cell states. That is how the manifold graph structure implicitly reflects cell developmental trajectories over longer distances although each single step is stochastic.
Palantir was published on Nature Biotechnology on Mar 21, 2019:
www.nature.com/articles/s41587-019-0068-4
The tool is available as a Python module at:
github.com/dpeerlab/Palantir/
REFERENCE:
1. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell, 2019. DOI: 10.1016/j.cell.2019.01.031
2. Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nature Neuroscience (2019). DOI: 10.1038/s41593-019-0349-8
Characterization of cell fate probabilities in single-cell data with Palantir. Nature Biotechnology (2019). DOI: https://doi.org/10.1038/s41587-019-0068-4
Gia
This is really helpful, thanks.