Highlights:
In every biology textbook, expression of the B cell receptor (BCR) defines B cells, and the T cell receptor (TCR) defines T cells. However, in 2019, Ahmed et al. discovered a strange cell population in Type I Diabetes that coexpresses the BCR and TCR, and key lineage markers of both B and T cells.
In this work, we compared the T cell populations in lung tumors vs normal lung tissue from all BioTuring lung cancer single cell data and surprisingly found that B Cell Receptor (BCR) light chain and heavy chain genes (specifically, IGKC and IGHA1) to be up-regulated in lung adenocarcinoma.
Comparing T cells populations in lung cancer datasets:
Lung cancer is the most common cancer in the world (Globocan 2018) (Fig.1). It can be broadly classified into two forms, small-cell lung carcinomas (SCLCs) and non-small-cell lung carcinomas (NSCLCs). In fact, lung adenocarcinoma is the most popular type of NSCLCs, which also accounts for 30-40% of lung cancer (Cersosimo RJ et al., 2002).
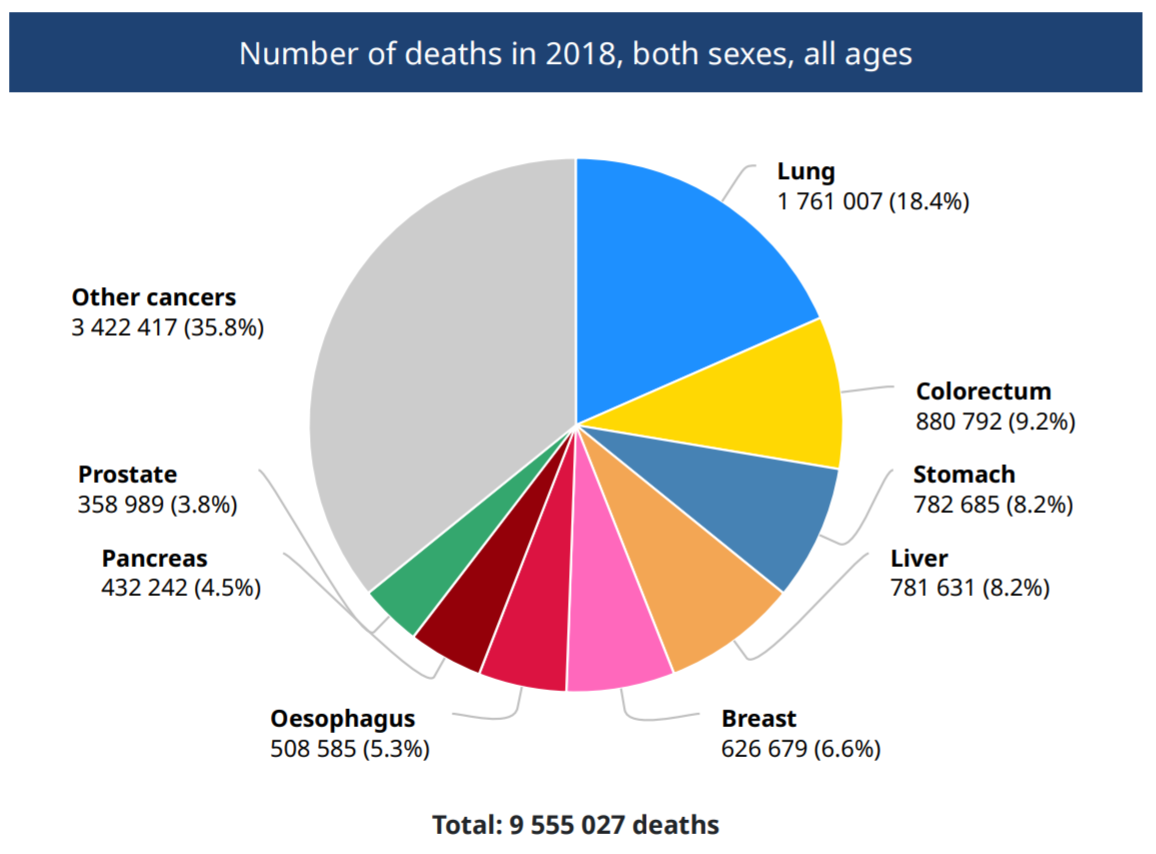
Figure 1. Number of deaths in 2018, both sexes, all ages (Globocan 2018)
To understand the immune cell dynamics in lung adenocarcinoma, we used BioTuring Browser Lung Cancer Single cell Database (consisting of more than 11.000.000 cells) to explore two lung adenocarcinoma datasets:
(1) Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma (Kim et al., 2020).
(2) Regenerative lineages and immune-mediated pruning in lung cancer metastasis (Laughney et al., 2020).
Results:
1.IGKC, IGLC2, IGHA1, and IGHG3 are up-regulated in T cells from lung tumor and brain metastasis in the dataset of Kim et al. (2020)
Using BioTuring Browser to find differentially expressed genes in T cells from the tumor vs. normal lung tissues, we found significant up-regulation of immunoglobulin genes in tumor-infiltrating T cells. These most significant genes included IGKC, IGLC2, IGHA1 and IGHG3 (Fig. 3, 4).
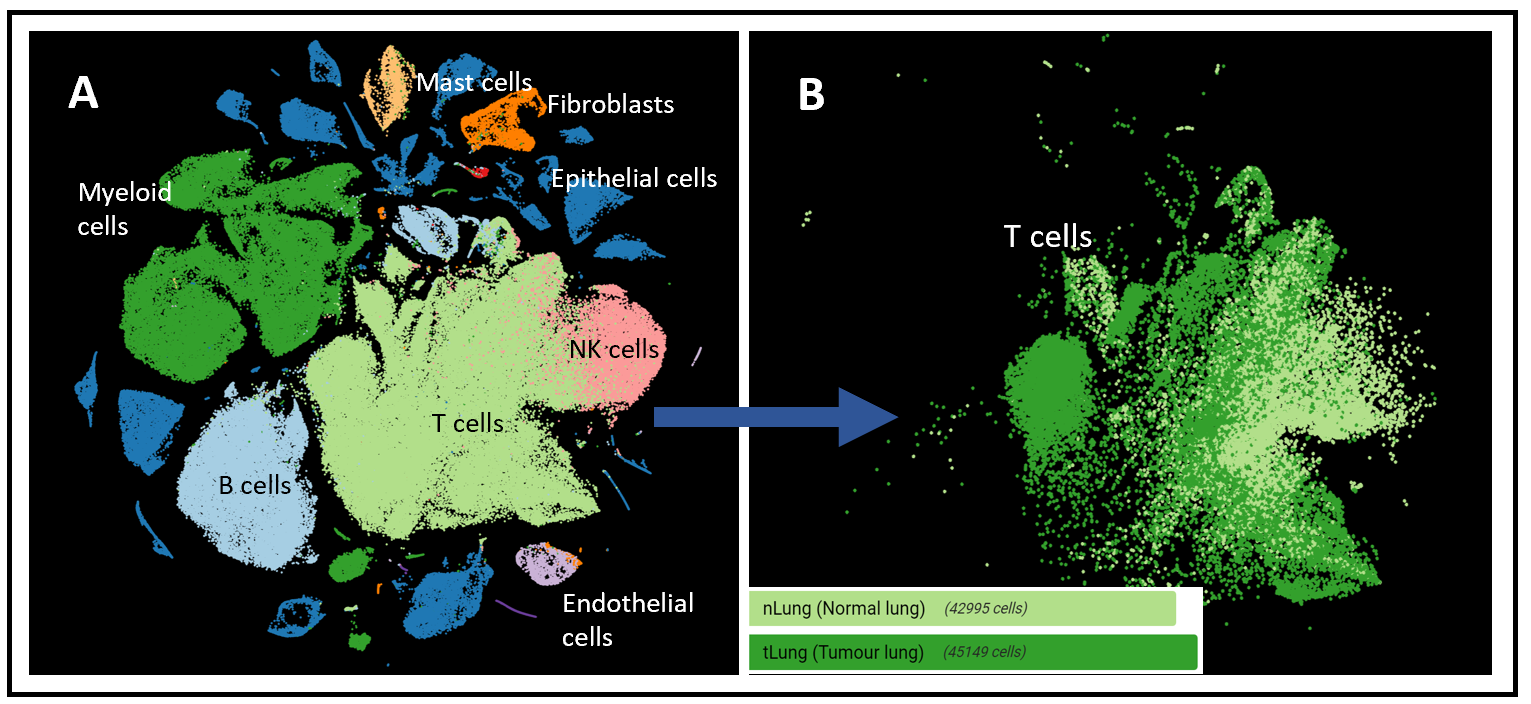
Figure 2.(A) t-SNE of all cell types (left) and (B) only the T cells (right) in the dataset of Kim et al. (2020), colored by normal lung (42.995 cells) and tumor lung (45.149 cells)
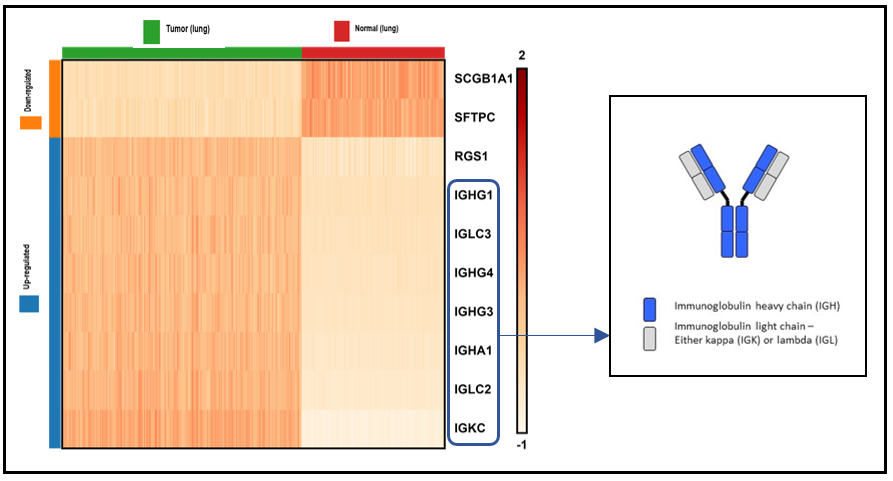
Figure 3. Heatmap of top 10 differentially expressed genes in T cells from the lung tumor vs. normal lung tissue
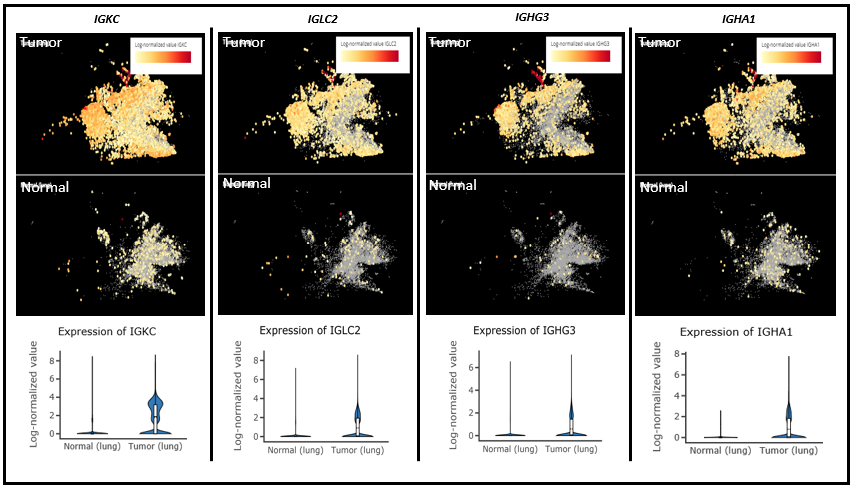
Figure 4. Top 4 Immunoglobulin genes up-regulated in Tumor T cells from left to right: IGKC, IGLC2, IGHG3 and IGHA1. Upper panel shows T cells from lung cancer while the lower panel shows T cells from normal lungs.
Meanwhile, IGKC, the most significant gene, was also highly up-regulated in the T cells of brain metastasis, compared with those in the normal lung tissues (Fig. 3). Our composition analysis for T-cells in tumors also showed that CD8+/CD4+ mixed T helper cells were the subtypes that exhibited the highest expression of IGKC (Fig. 5).
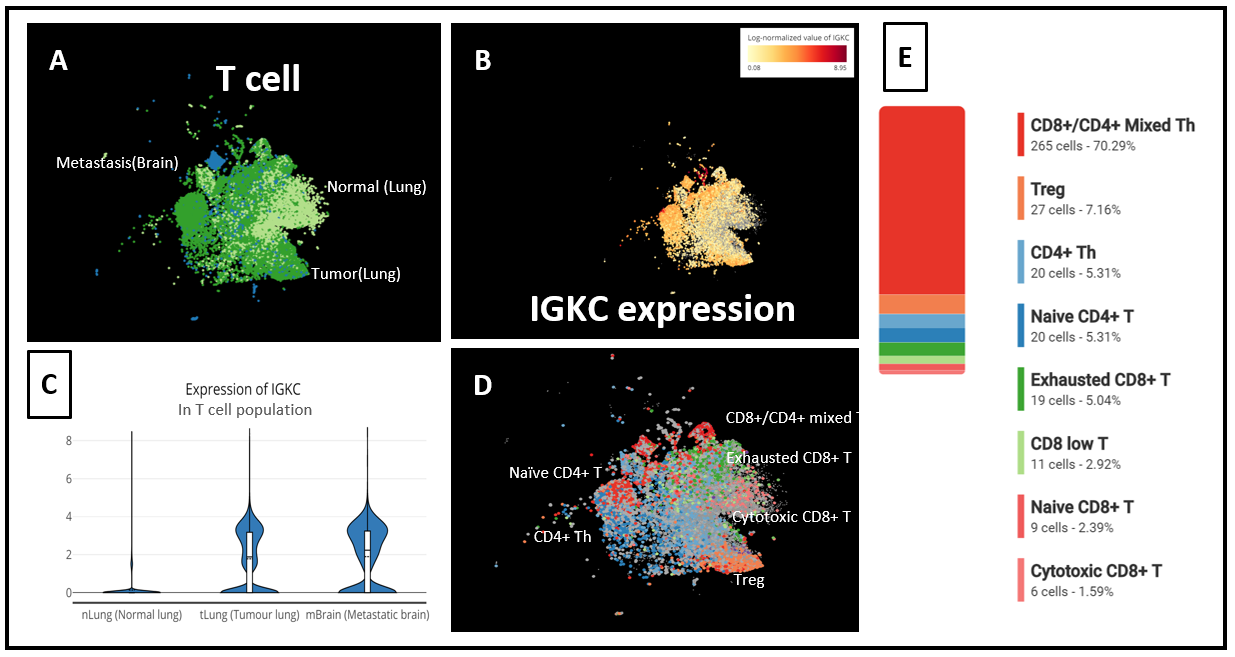
Figure 5. (A) t-SNE of T cells from 3 groups: Brain Metastasis, Normal (Lung) and Tumor (lung), (B) IGKC expression level in T cells, (C) IGKC expression across T cells from Brain Metastasis, Normal (Lung) and Tumor (lung), (D) T cell subtypes identified in the study, (E) Composition analysis showed that top IGKC expressors in Tumor (lung) were mainly CD8+/CD4+ mixed T helper cells
2.IGKC, IGLC2, IGLC3 and IGHA1 expression gradually increased in T cells from normal tissues, to tumor, to metastasis in the dataset of Laughney et al. (2020)
In the dataset of Laughney and colleagues (2020), we also ran differential expression analysis on CD3+ cell populations from the tumor vs. normal lung tissue. And immunoglobulin genes is also up-regulated in the T cells from tumor and metastasis groups, compared with normal lung tissues (Fig. 6 A,B,C).
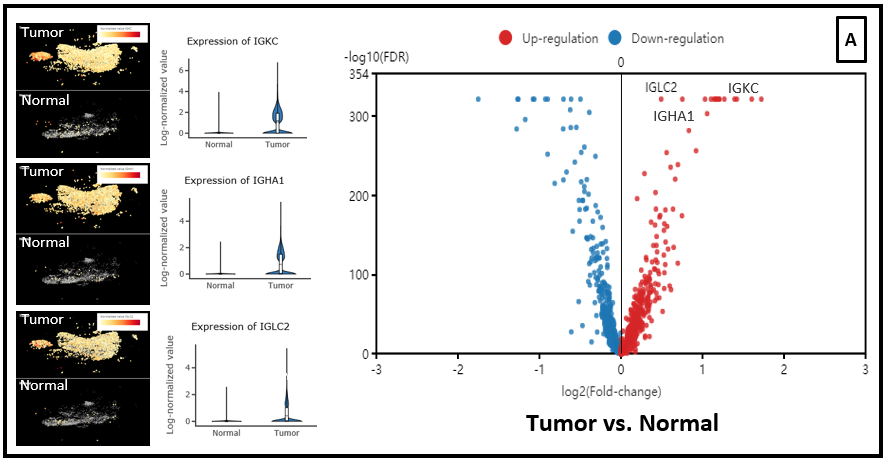
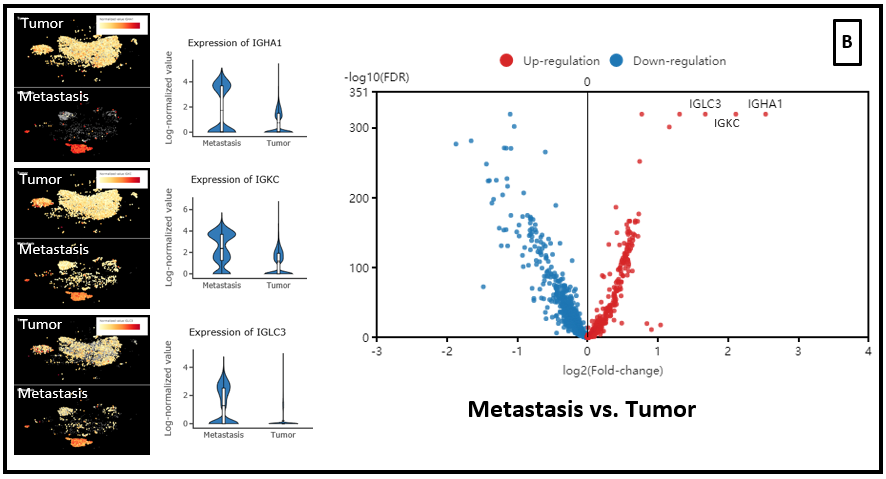
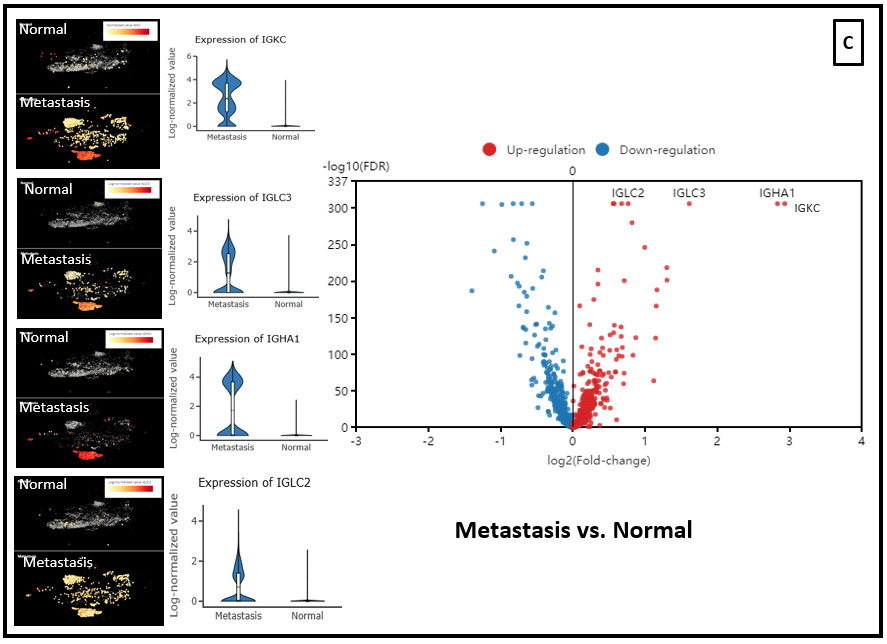
Figure 6. Differential expression analysis between T cell population of (A) Tumor vs. Normal groups, (B) Metastasis vs. Tumor, (C) Metastasis vs. Normal
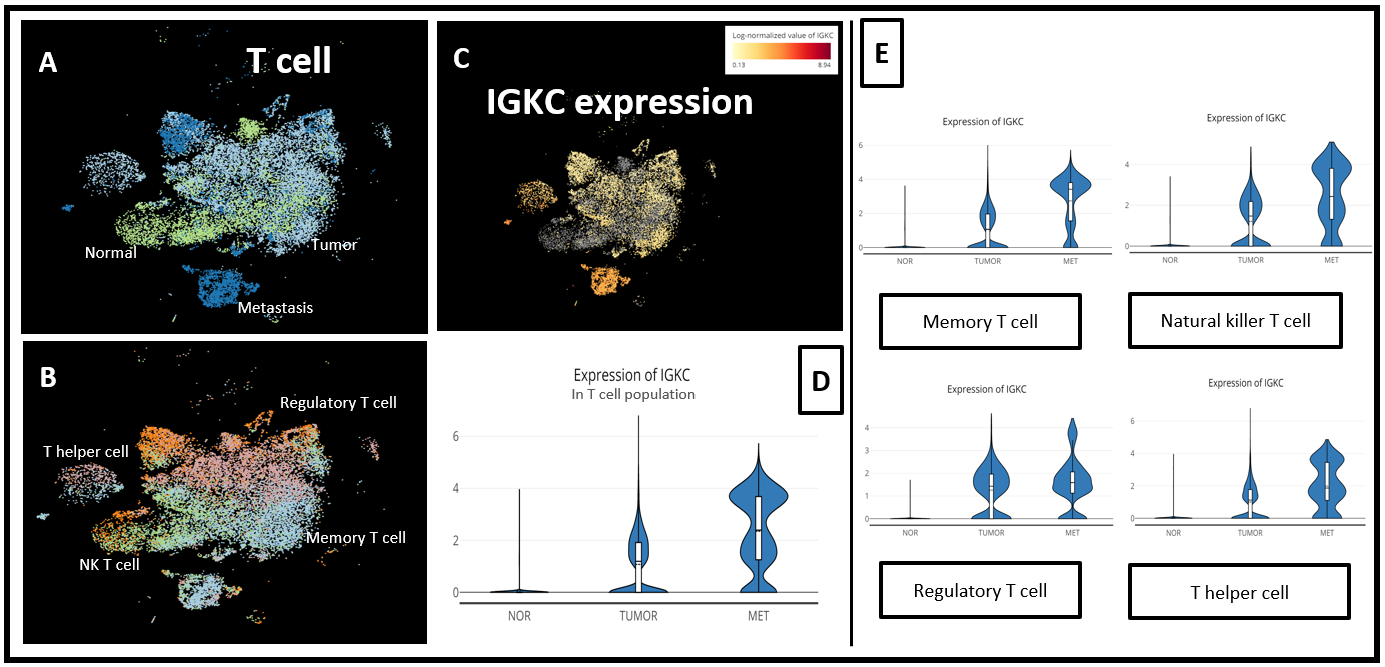
Figure 7. (A) t-SNE of the T cell population of 3 groups: Metastasis, Normal and Tumor, and (B) all T cell subtypes in 3 groups; (C) t-SNE of IGKC expression and (D) comparison of IGKC expression level across 3 conditions in the entire T cell population and in (E) each T cell subtype.
Interestingly, in this dataset, IGKC was gradually up-regulated in T cells from normal tissues to tumor to metastasis. In particular, this appeared in the memory T cell, NK T cell, and T helper cell (Fig. 7).
Discussion
In 2019, Ahmed et al. discovered a dual expresser cell population in Type 1 Diabetes which expresses genes specific to both B and T cell lineages and also their receptors. These dual expresser cells are marked by BCR (CD79a, CD79b)/(CD19 and CD20) with light/heavy chains and TCR (CD3G, CD3E, CD3D, and CD247) (Ahmed et al.,2019).
What is the role of these dual expresser cells? In type I diabetes, HLA-DQ8 is known to mistakenly present insulin to T cells in such a way that stimulates T cells to attack the pancreatic β cells, which produce insulin. Interestingly, these dual expresser cells have a single BCR clonotype (IGHV), which produces a peptide that is bound tightly to the HLA-DQ8 molecule. This stimulated T helper cells from Type 1 Diabete patients to proliferate and to secrete proinflammatory cytokines. The T helper cells then go on to direct certain killer T cells to eliminate β cells of the pancreas (Ahmed et al., 2019).
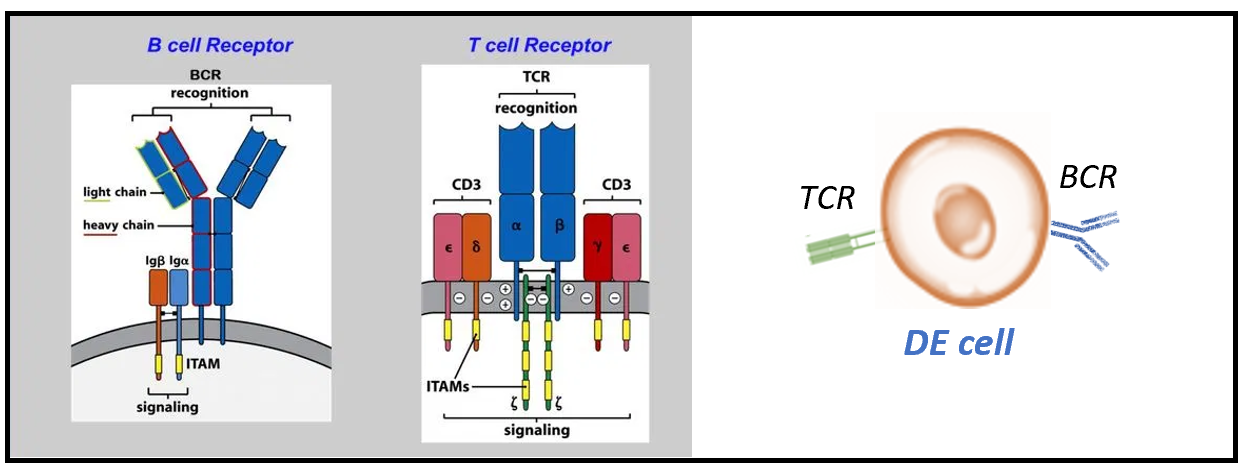
Figure 8. Dual expresser (DE) cells comprise of both BCRs and TCRs in T1D (Ahmed et al.,2019)
Does it mean the immunoglobulin up-regulation in tumor-infiltrating T cells triggers the immune response? And against what?
To answer this, we need more data, and research across other cancer types, to understand whether this immunoglobulin up-regulation also happens consistently.
For detailed instructions on how to reproduce the analysis, check out our video below:
References
- Cersosimo RJ. Lung cancer: a review. Am J Health Syst Pharm. 2002;59(7):611-642. doi:10.1093/ajhp/59.7.611
- Neeve SC, Robinson BW, Fear VS. The role and therapeutic implications of T cells in cancer of the lung. Clin Transl Immunology. 2019;8(8):e1076. Published 2019 Aug 28. doi:10.1002/cti2.1076
- Rizwan Ahmed et al. A Public BCR Present in a Unique Dual-Receptor-Expressing Lymphocyte from Type 1 Diabetes Patients Encodes a Potent T Cell Autoantigen, 2019, Cell. Volume 177, Issue 6,1583-1599.e16. https://doi.org/10.1016/j.cell.2019.05.007
- Nayoung Kim et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. 2020. Nat Commun 11, 2285 (2020). https://doi.org/10.1038/s41467-020-16164-1
- Laughney A.M., Hu, J., Campbell, N.R. et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. 2020. Nat Med 26, 259–269. https://doi.org/10.1038/s41591-019-0750-6
- Lehuen, A., Diana, J., Zaccone, P. et al. Immune cell crosstalk in type 1 diabetes. 2010. Nat Rev Immunol 10, 501–513. https://doi.org/10.1038/nri2787
This article is for research purposes only. Please do not use this for clinical purposes to any extent.