Single-cell sequencing technologies have brought up an unprecedented level of resolution to omics studies with extremely detailed views into individual cells’ expression patterns. With the emergence of drop-based sequencing methods, the resolution now even comes with scale and efficiency. In a single experiment, scientists can now profile hundreds of thousands of cells, or even 2 million cells at a time (Cao et. al., 2019), and generate a huge amount of single-cell data, providing valuable inputs for biological research and breakthroughs.
However, huge amounts of single-cell data generated are posing major challenges for hosting, accessing, visualization and analysis. Although multiple single-cell databases are available (eg.: Gene Expression Omnibus by NCBI, EBI, Human Cell Atlas, etc.), single-cell sequencing datasets are currently provided in different formats. Given the increasing size of the data, it is extremely challenging to explore many single-cell datasets at a time because of such variability in formats, and expensive computational resources to process. We are lucky to have some nice visualization portals at hand, thanks to the authors of the data, yet these portals are separate from one another, making it uneasy to combine multiple single-cell datasets for meta-analysis.
To address the challenges, we developed a modern single-cell database and standardized analytics dashboard in BioTuring Single-cell Browser, enabling any biologists to easily access, reanalyze, combine, and discover valuable stories from published single-cell datasets on their laptop.
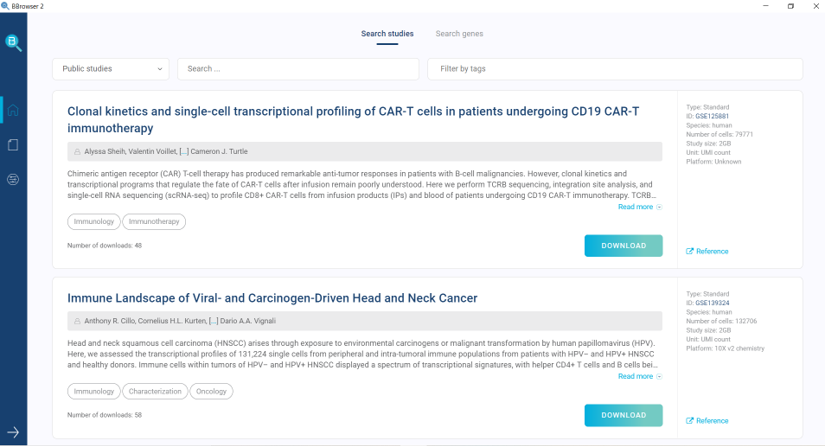
The single-cell database in BioTuring Single-cell Browser
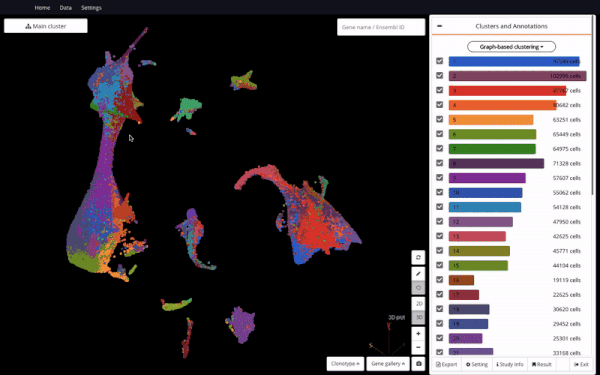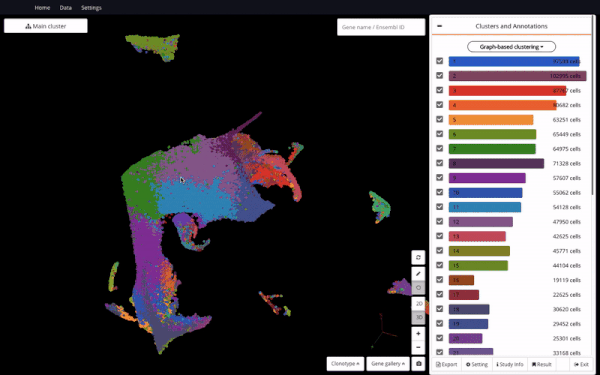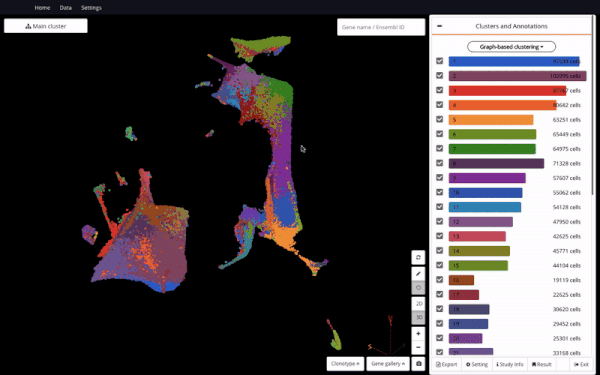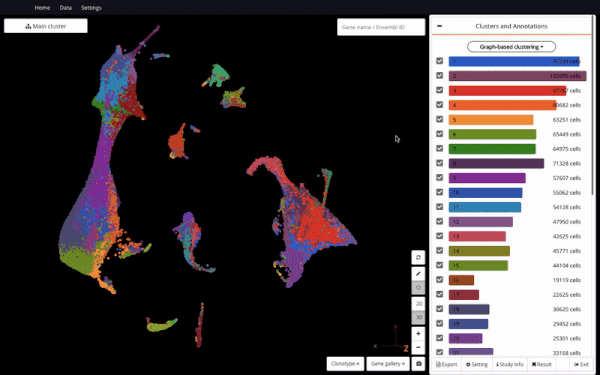
3D UMAP of 1.3 million cells from the mouse organogenesis cell atlas (MOCA) (Cao et al., 2019) in BioTuring Single-cell Browser
A curated single-cell database of 5.9 million cells, ready for downstream analyses
Our curated single-cell database in BioTuring Single-cell Browser is currently hosting more than 5.9 million cells from 29 diseases and more than 150 latest high-impact single-cell studies, including the mouse organogenesis cell atlas by Cao and colleagues (2019), mouse retinal cell atlas (Clark et al., 2019) and the liver cell atlas (MacParland et al., 2018). The datasets are increasing exponentially in number, all ready for exploring in a few clicks, with interactive visualizations, and state-of-the-art analytics.
List of indexed datasets (as of Oct, 2019)
Total Single-cell RNA-seq studies in BBrowser
| Title | ID | Specie | # cells |
|---|---|---|---|
| A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells | Singer2016 | mouse | 1061 |
| Massively parallel single-nucleus RNA-seq with DroNc-seq | habib2017_2 | human | 14963 |
| Shared and distinct transcriptomic cell types across neocortical areas | tasic2018 | mouse | 11430 |
| Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. | GSE123139 | human | 64967 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - EntoPeduncular | dropviz.EntoPeduncular | mouse | 27927 |
| GSE121645_age - Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes | GSE121645_age | mouse | 85254 |
| A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex | zhong2018 | human | 2394 |
| Neutrophils escort circulating tumour cells to enable cell cycle progression | GSE109761_hu | human | 327 |
| Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type | boldog2018 | human | 914 |
| Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment - BC09-01-vdj | bc09_1_vdj | human | 7096 |
| Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing | Zheng2017 | human | 5063 |
| Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade | Ishizuka2018 | mouse | 8838 |
| Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets | macosko2015 | mouse | 49300 |
| Longitudinal single-cell RNA sequencing of patient-derived primary cells reveals drug-induced infidelity in stem cell hierarchy | Sharma2018 | human | 1302 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - Cortex_noRep5_POSTERIORonly | dropviz.Cortex_noRep5_POSTERIORonly | mouse | 115990 |
| Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma | darmanis2017 | human | 3588 |
| GSE121645_saline - Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes | GSE121645_saline | mouse | 13309 |
| Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations | sonya2018 | human | 8444 |
| Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes | romanov2017 | mouse | 3077 |
| An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics | Angelidis2018 | mouse | 14578 |
| Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer | GSE103322 | mouse | 5902 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - GlobusPallidus | dropviz.GlobusPallidus | mouse | 86453 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - Striatum | dropviz.Striatum | mouse | 90470 |
| Composition of the Colonic Mesenchyme and the Nature of its Plasticity in Inflammatory Bowel Disease | GSE114374 | human | 9271 |
| Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing | xinyi2018 | human | 11840 |
| Neutrophils escort circulating tumour cells to enable cell cycle progression | GSE109761_mm | mouse | 109 |
| Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors | young2018 | human | 95682 |
| Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification | MouseRetina | mouse | 102830 |
| Single-cell profiling of the myeloid landscape identifies cell subsets with distinct fates during neuroinflammation | GSE118948 | mouse | 4509 |
| Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type | boldog2018_2 | mouse | 1679 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - SubstantiaNigra | dropviz.SubstantiaNigra | mouse | 66346 |
| Temporal tracking of microglia activation in neurodegeneration at single-cell resolution | mathys2017 | mouse | 2208 |
| Single-cell reconstruction of the early maternal--fetal interface in humans | ventotormo2018 | human | 64734 |
| Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq | zeisel2015 | mouse | 3488 |
| Massively parallel single-nucleus RNA-seq with DroNc-seq | habib2017 | mouse | 13313 |
| The bone marrow microenvironment at single-cell resolution | GSE108892-GSE108891 | mouse | 20094 |
| Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain | lake2016 | human | 3042 |
| Classes and continua of hippocampal CA1 inhibitory neurons revealed by single-cell transcriptomics | harris2018 | mouse | 3662 |
| A molecular atlas of cell types and zonation in the brain vasculature | vanlandewijck2018 | mouse | 3186 |
| Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment - BC01-08 | GSE114725 | human | 28690 |
| Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment - BC09-11 | GSE114727 | human | 28690 |
| The single-cell transcriptional landscape of mammalian organogenesis | MOCA_2M | mouse | 1331984 |
| The bone marrow microenvironment at single-cell resolution | GSE108892-GSE118436 | mouse | 23029 |
| Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma | Adit2018 | human | 40065 |
| Dissecting cell-type composition and activity-dependent transcriptional state in mammalian brains by massively parallel single-nucleus RNA-seq | hu2017 | mouse | 18194 |
| A unique microglia type associated with restricting development of Alzheimer’s disease | Hadas2017 | human | 37248 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - Cerebellum | dropviz.Cerebellum_ALT | mouse | 59309 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - Thalamus | dropviz.Thalamus | mouse | 124239 |
| Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain - Hippocampus | dropviz.Hippocampus | mouse | 131953 |
| GSE121645_percoll - Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes | GSE121645_percoll | mouse | 15281 |
| Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity | GSE116256 | human | 15281 |
| Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris (FACS) | TabulaMuris_facs | mouse | 40494 |
| Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris (droplet) | TabulaMuris_droplet | mouse | 40494 |
| Colonic epithelial cell diversity in health and inflammatory bowel disease | GSE116222 | human | 11167 |
| Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response | GSE109564-GSE114156 | human | 8784 |
| Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics | GSE118184 | human | 89333 |
| Mapping the Mouse Cell Atlas by Microwell-Seq | MCA | mouse | 378824 |
| Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species | GSE127465_human | human | 53179 |
| Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species | GSE127465_mouse | mouse | 15915 |
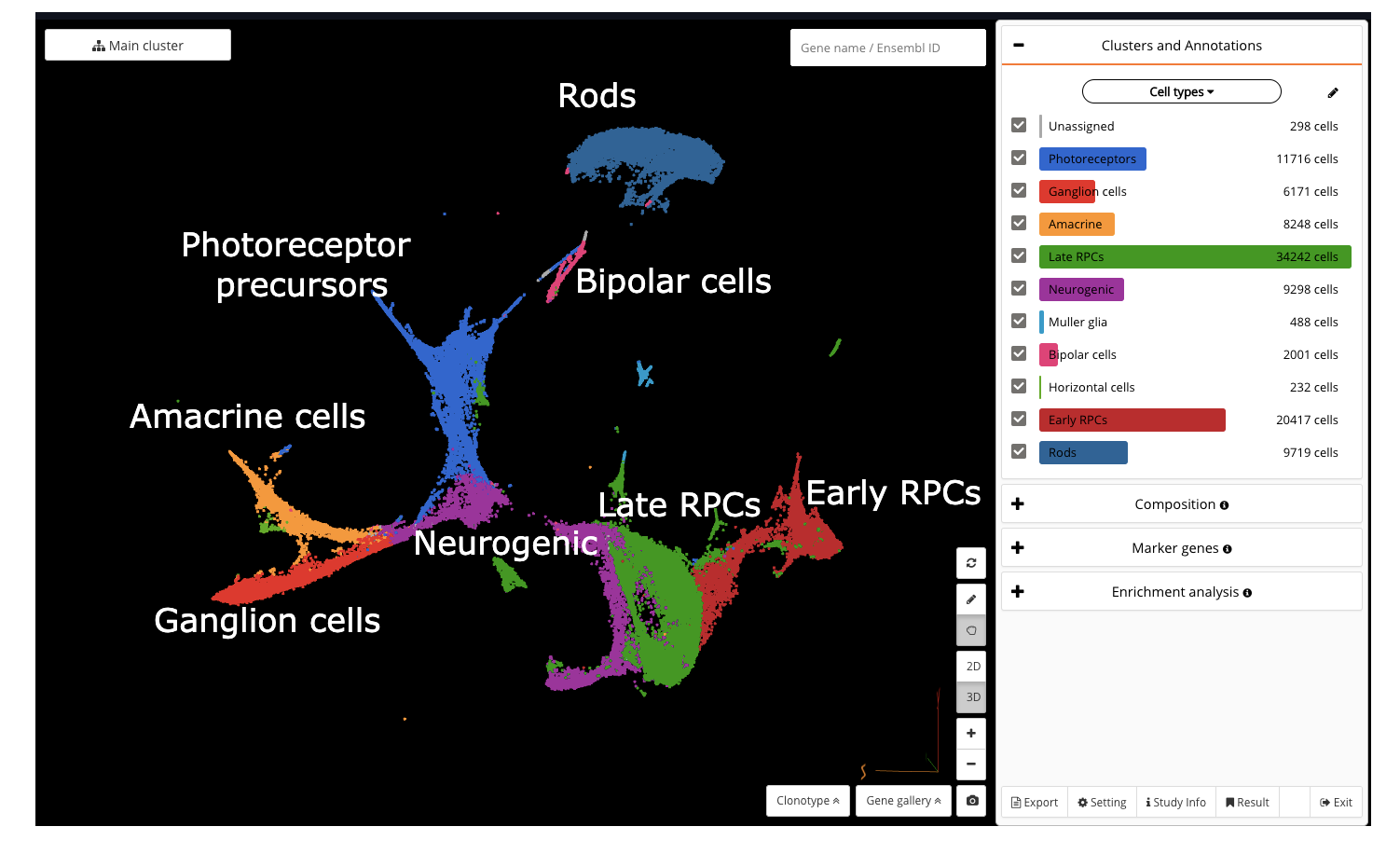
UMAP of mouse retinal cell atlas (Clark et al., 2019) in BioTuring Single-cell Browser. See how we reanalyzed the dataset and explored a hybrid population expressing rod and cone markers here.
What can you do in BioTuring Single-cell Browser?
Here’s to name major analyses available for published single-cell data in BioTuring Browser:
- Query gene expression and search for similar populations across the database (Cell Search)
- Interactively visualize up to 1.3 million cells in 2D/3D t-SNE and UMAP
- Find marker genes and predict cell types in real time
- Explore sub-clusters of any cell types
- Study compositional changes across conditions
- Find differentially expressed genes
- Pair clonotype data from TCR repertoire sequencing with expression data
- CITE-Seq data analysis
- Combine multiple published single-cell datasets or pair published data with your in-house data
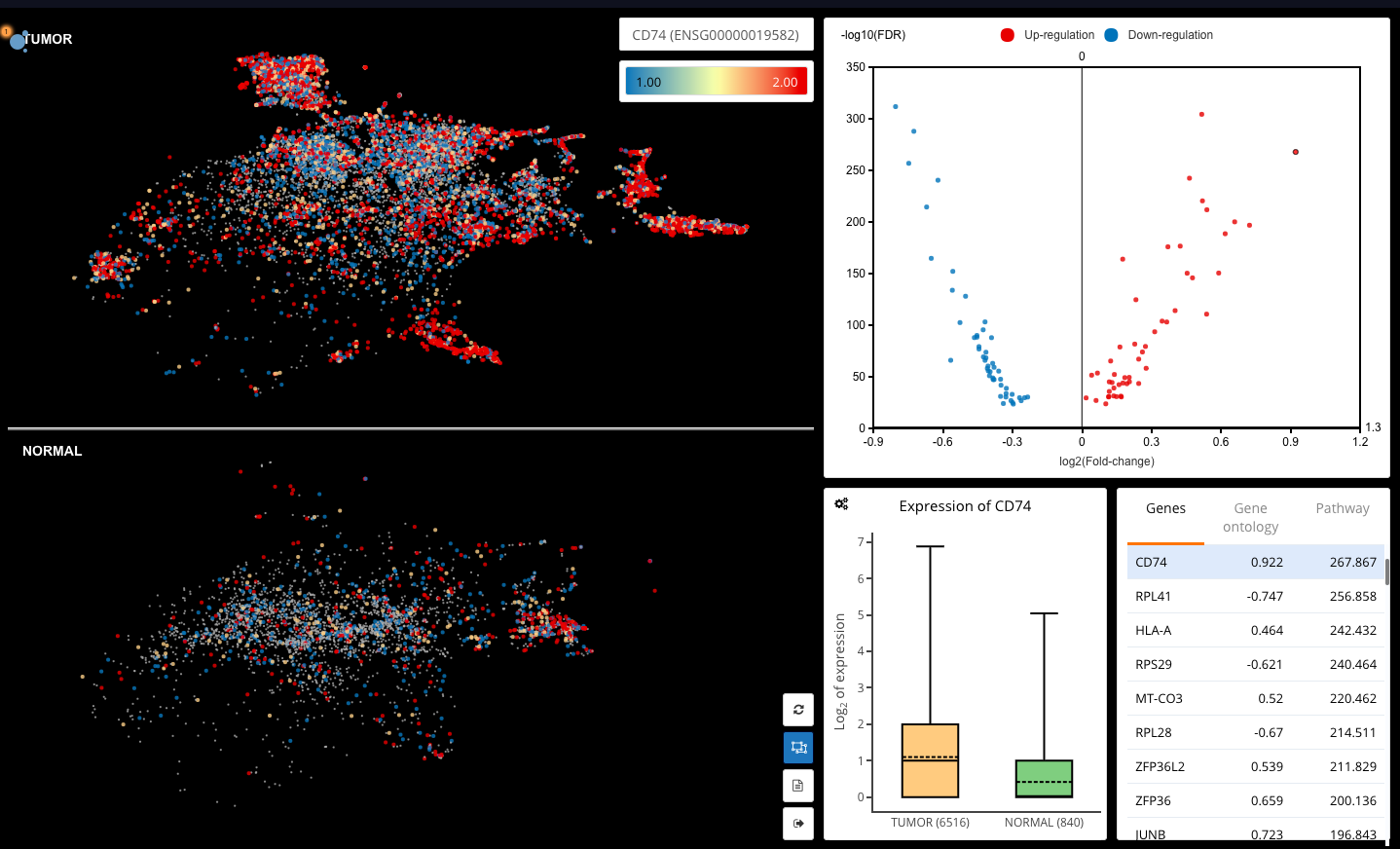
Differential expression analysis for T cells in tumor and normal breast tissues (Azizi et al., 2018)
Explore now
BioTuring Single-cell Browser is now available at bioturing.com/product/bbrowser for Windows, MacOS, Ubuntu and Centos 7.
For more video tutorials:
https://www.youtube.com/watch?v=w7wu69HAHEw&list=PLgssfInwsHRbhV3FcCi4c99mzpVRWkrLZ
References:
Cao, Junyue, et al. "The single-cell transcriptional landscape of mammalian organogenesis." Nature 566.7745 (2019): 496.
Clark, Brian S., et al. "Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification." Neuron (2019).
MacParland, Sonya A., et al. "Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations." Nature communications 9.1 (2018): 4383.
Azizi, Elham, et al. "Single-cell map of diverse immune phenotypes in the breast tumor microenvironment." Cell 174.5 (2018): 1293-1308.