B cells are like key ‘defenders’ in a soccer match. They play a critical role in supporting T cells, the ‘strikers’, in their battle against diseases, by producing antibodies to block specific antigens. Yet there are different types of defenders. Some form the memory that remembers the same pathogen for faster antibody production in future infections (memory B cells). Some secrete the antibodies in large amounts (plasma cells). Some are reserved and ready to fight when they are called (naive B cells). Even in complex diseases such as lung cancer, B cell subsets are much more diverse (Stankovic et al., 2019; van de Veen et al., 2020).
Fig.1. Shared and unique B cell subsets in 6 lung datasets curated from BioTuring single-cell database.
To explore the complex landscape of B cell subsets, we subcluster the B cell populations in 6 human lung single-cell datasets in BBrowser (Maynard et al.,2020; Kim et al., 2020; Laughney et al., 2020; Lambrechts et al., 2018; Adams et al., 2020; Morse et al., 2019). Our revisit detects 5 subsets that are shared across the datasets and 3 unique subsets in each.
1. Shared B cell subsets
-
BANK1+ B cells
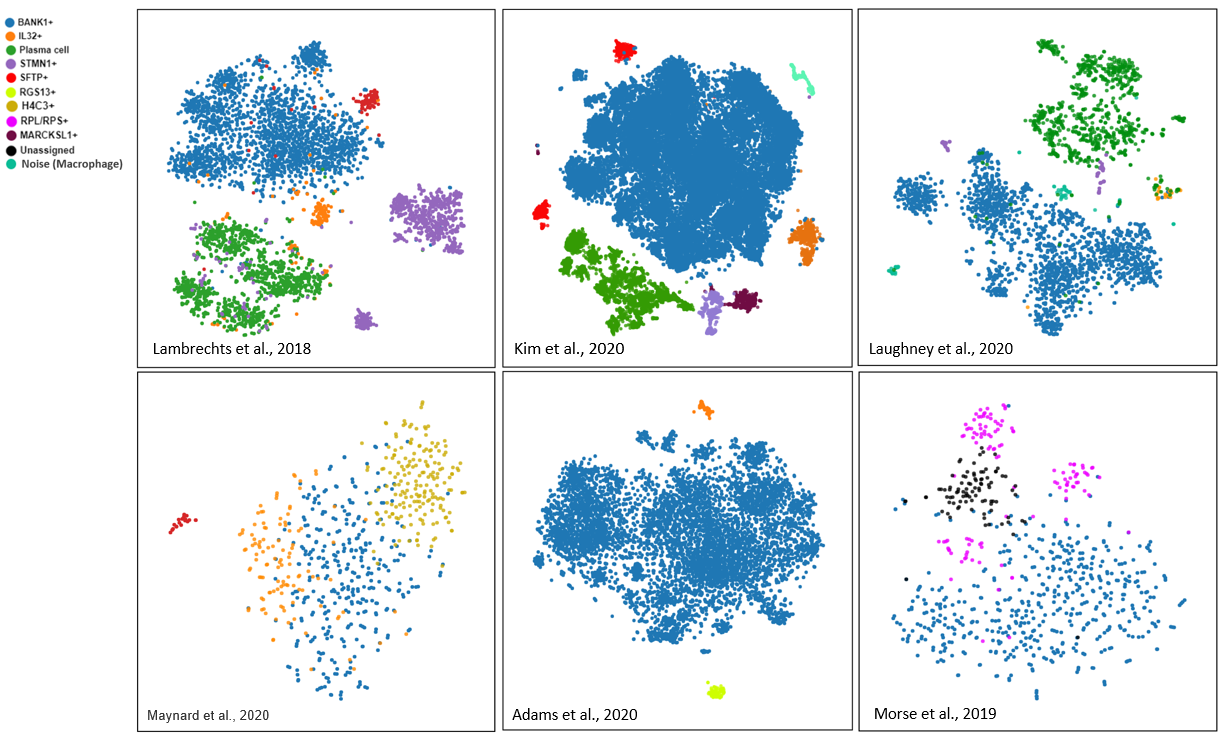
We consistently found BANK1+ B cell clusters in all 6 studies (Maynard et al.,2020; Kim et al., 2020; Laughney et al., 2020; Lambrechts et al., 2018; Adams et al., 2020; Morse et al., 2019). BANK1 (B cell scaffold protein with ankyrin repeats 1) promotes tyrosine phosphorylation of the IP3 receptor to modulate B cell antigen receptor (BCR), inducing calcium mobilization (Yokohama et al., 2002). The cluster also upregulates HLA-DRA, MS4A1, CD37, and HLA-DPB1 in some lung cancer datasets (Lambrechts et al., 2018; Kim et al., 2020; Laughney et al., 2020).
Fig.2. BANK1+ B cell subsets in 6 lung datasets.
-
Plasma cells
Plasma cell clusters appear in 3 datasets (Lambrechts et al., 2018; Kim et al., 2020; Laughney et al., 2020) with unique markers of Ig genes such as JCHAIN, IGHA1, IGHA2, IGLC2, IGLC3, etc.
Fig.3. Plasma cells with some Ig genes’ expression in 3 lung datasets.
-
IL32+ CD3+ B cells
Unexpectedly, we found a B cell subset that expressed IL32 and CD3 in 5 datasets (Maynard et al.,2020; Kim et al., 2020; Laughney et al., 2020; Lambrechts et al., 2018; Adams et al., 2020). It is interesting that the subset expresses B cell markers (e.g. CD79A, CD79B, CD19, CD20) together with T cell markers (e.g. CD3D, CD3E) and IL32. Interleukin 32 (IL-32) is a proinflammatory cytokine that can make immune cells to secrete inflammatory cytokines, like tumor necrosis factor-alpha (TNF-α) and IL-6. A previous study found that IL-32 expression also appeared in B cells (Smith et al., 2012). Furthermore, these IL32+ clusters also upregulate a set of genes, such as CCL5, TRAC, FYB, and CD2 (Maynard et al.,2020; Kim et al., 2020; Laughney et al., 2020; Lambrechts et al., 2018; Adams et al., 2020).
Fig.4. IL32+ B cells
-
SFTPC+ B cells
We also found SFTPC+ B cells in 3 studies (Maynard et al.,2020; Kim et al., 2020; Lambrechts et al., 2018). In these datasets, SFTPC+ clusters also upregulate NAPSA, RPL/RPS genes, and SFTPA1/A2. However, no reports have shown SFTPC positive expression on B cells.
Fig.5. SFTPC+ B cells in 3 datasets.
2. Specific subsets in each study
In addition, we spotted several B cell subsets specific to each dataset, like RGS13+ B cells, H4C3+ B cells, and MARCKSL1+ B cells (Maynard et al.,2020; Kim et al., 2020; Adams et al., 2020 respectively). There are some biological meanings related to these 3 genes:
- RGS13 is a member of the regulator of the G protein signaling (RGS) family. It can inhibit G protein-coupled receptor signaling in B cells and mast cells (Iwaki et al., 2011). This cluster also highly expresses CCDC144B, CCDC144CP, LRMP, LPP, etc.
- H4C3, known as H4 clustered histone 3, encodes basic nuclear proteins that contribute to the nucleosome structure of the chromosomal fiber, but few reports have been published related to this gene in B cells. H4C3+ cluster also expresses COX7A2, LSM7, COMMD6, etc.
- MARCKSL1, by regulating epithelial-mesenchymal transition, can promote the progression of lung adenocarcinoma (Liang et al., 2020). This gene also appears in the transitional 1 immature and mature B lymphocytes in response to B-cell antigen receptor triggering (Murn et al., 2009). ACTG1, ACTB, HMGN1,… are also highly expressed in the MARCKSL1+ cluster.
Fig.6. Specific B cell subsets in 3 datasets.
Above are some shared and unique B cell subsets in 6 curated lung single-cell datasets in BioTuring Browser. We have not fully interpreted their biological meanings yet, but hope this work helps to sketch a picture of how complex B cell subsets are in the lungs. Let us know if you have found the same subsets!
Copyright 2021. BioTuring Inc. This article is for research purposes only.
REFERENCES
- Stankovic, B., Bjørhovde, H. A. K., Skarshaug, R., Aamodt, H., Frafjord, A., Müller, E., . . . Corthay, A. (2019). Immune Cell Composition in Human Non-small Cell Lung Cancer. Frontiers in Immunology, 9, 3101. https://doi.org/10.3389/fimmu.2018.03101
- van de Veen, W., Globinska, A., Jansen, K., Straumann, A., Kubo, T., Verschoor, D., . . . Akdis, M. (2020). A novel proangiogenic B cell subset is increased in cancer and chronic inflammation. Science Advances, 6(20), raaz 3559. doi:10.1126/sciadv.aaz3559
- Adams, T. S., Schupp, J. C., Poli, S., Ayaub, E. A., Neumark, N., Ahangari, F., . . . Kaminski, N. (2020). Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Science Advances, 6(28), eaba1983. doi:10.1126/sciadv.aba1983
- Iwaki, S., Lu, Y., Xie, Z., & Druey, K. M. (2011). p53 Negatively Regulates RGS13 Protein Expression in Immune Cells Journal of Biological Chemistry, 286(25), 22219-22226. doi:10.1074/jbc.M111.228924
- Kim, N., Kim, H. K., Lee, K., Hong, Y., Cho, J. H., Choi, J. W., . . . Lee, H.-O. (2020). Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nature Communications, 11(1), 2285. doi:10.1038/s41467-020-16164-1
- Lambrechts, D., Wauters, E., Boeckx, B., Aibar, S., Nittner, D., Burton, O., . . . Thienpont, B. (2018). Phenotype molding of stromal cells in the lung tumor microenvironment. Nature Medicine, 24(8), 1277-1289. doi:10.1038/s41591-018-0096-5
- Laughney, A. M., Hu, J., Campbell, N. R., Bakhoum, S. F., Setty, M., Lavallée, V. P., . . . Massagué, J. (2020). Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med, 26(2), 259-269. doi:10.1038/s41591-019-0750-6
- Li, Z., Wang, S., Huo, X., Yu, H., Lu, J., Zhang, S., . . . Chen, Z. (2018). Cystatin C Expression is Promoted by VEGFA Blocking, With Inhibitory Effects on Endothelial Cell Angiogenic Functions Including Proliferation, Migration, and Chorioallantoic Membrane Angiogenesis. Journal of the American Heart Association, 7(21), e009167. doi:10.1161/JAHA.118.009167
- Liang, W., Gao, R., Yang, M., Wang, X., Cheng, K., Shi, X., . . . Yu, X. (2020). MARCKSL1 promotes the proliferation, migration and invasion of lung adenocarcinoma cells. Oncol Lett, 19(3), 2272-2280. doi:10.3892/ol.2020.11313
- Maynard, A., McCoach, C. E., Rotow, J. K., Harris, L., Haderk, F., Kerr, D. L., . . . Bivona, T. G. (2020). Therapy-Induced Evolution of Human Lung Cancer Revealed by Single-Cell RNA Sequencing. Cell, 182(5), 1232-1251.e1222. https://doi.org/10.1016/j.cell.2020.07.017
- Morse, C., Tabib, T., Sembrat, J., Buschur, K. L., Bittar, H. T., Valenzi, E., . . . Lafyatis, R. (2019). Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J, 54(2). doi:10.1183/13993003.02441-2018
- Murn, J., Mlinaric-Rascan, I., Vaigot, P., Alibert, O., Frouin, V., & Gidrol, X. (2009). A Myc-regulated transcriptional network controls B-cell fate in response to BCR triggering. BMC Genomics, 10, 323-323. doi:10.1186/1471-2164-10-323
- Smith, A. J., Toledo, C. M., Wietgrefe, S. W., Duan, L., Schacker, T. W., Reilly, C. S., & Haase, A. T. (2011). The immunosuppressive role of IL-32 in lymphatic tissue during HIV-1 infection. Journal of immunology (Baltimore, Md. : 1950), 186(11), 6576-6584. doi:10.4049/jimmunol.1100277
- Yang, J., Ren, J., Yang, Y., Sun, J., Zhou, X., Zheng, S., . . . Kong, N. (2018). BANK1 alters B cell responses and influences the interactions between B cells and induced T regulatory cells in mice with collagen-induced arthritis. Arthritis research & therapy, 20(1), 9-9. doi:10.1186/s13075-017-1503-x