Multiple studies have shown smoking’s effects on the human bronchi. However, few have characterized its precise impact at cellular resolution. Published recently on Science Advances, a study by Duclos and colleagues has employed single-cell RNA sequencing into exploring the cellular changes in the human bronchial epithelium between current and never smokers, discovering a compositional repolarization, a detoxification program and a novel ciliated cell population present in current smokers.
This interesting single-cell data set has been indexed into BioTuring Single-cell Browser (or BBrowser) for easy access by the scientific community. In this episode of BioTuring Cellpedia, let’s check out some general information about the dataset and how you can reproduce such findings in BBrowser.
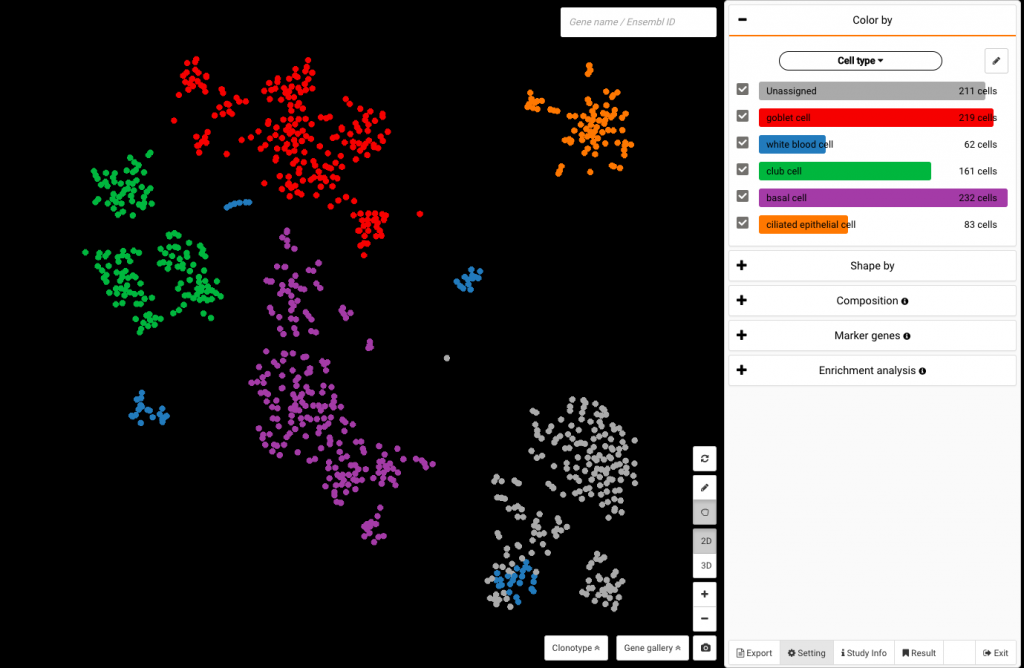
The data set by Duclos and colleagues visualized in BBrowser, colored by authors’ cell type annotations
Study info
*Accession ID in BBrowser: GSE131391
*Library construction method: CEL-Seq (modified)
*Type of data: UMI count data
*Species: human
*Disease: none
*Sampling site: right mainstem bronchus
*Cell types:
- Category: epithelial cells, blood cells
- Cell types: ciliated epithelial cells, goblet cells, club cells, basal cells, white blood cells
*Samples:
- 6 healthy current smokers
- 6 healthy never smokers
*Applications: lung cancer, lung disease, respiratory disease
*Keywords: single-cell RNA sequencing, single-cell data, lung disease, bronchial epithelium, bronchus, smoking-induced, epithelial cells
*Key findings:
- An expansion of goblet cells and depletion of club cells in the airways of smokers
- Smokers’ ciliated cells express a specific set of detoxification genes in response to smoke exposure.
- Discovery of an uncharacterized population named peri-goblet cells associated with smoking-induced goblet cell hyperplasia (GCH)
How to explore the data?
Access it
Open BBrowser and simply search for the data set by its title “Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution”, or by its ID “GSE131391”. Click the Download button when you have found it and click Explore when the download is complete.
Explore the cell types
Once opened, the data will be visualized in a t-SNE plot, and automatically colored by graph-based clustering results. To view the data with cell type labels retrieved from the authors, click the drop-down arrow at the Color by panel and select Cell type at Your annotation part.
To view BioTuring’s standardized cell type annotations with ontology numbers, select BioTuring – Cell type under the same panel.
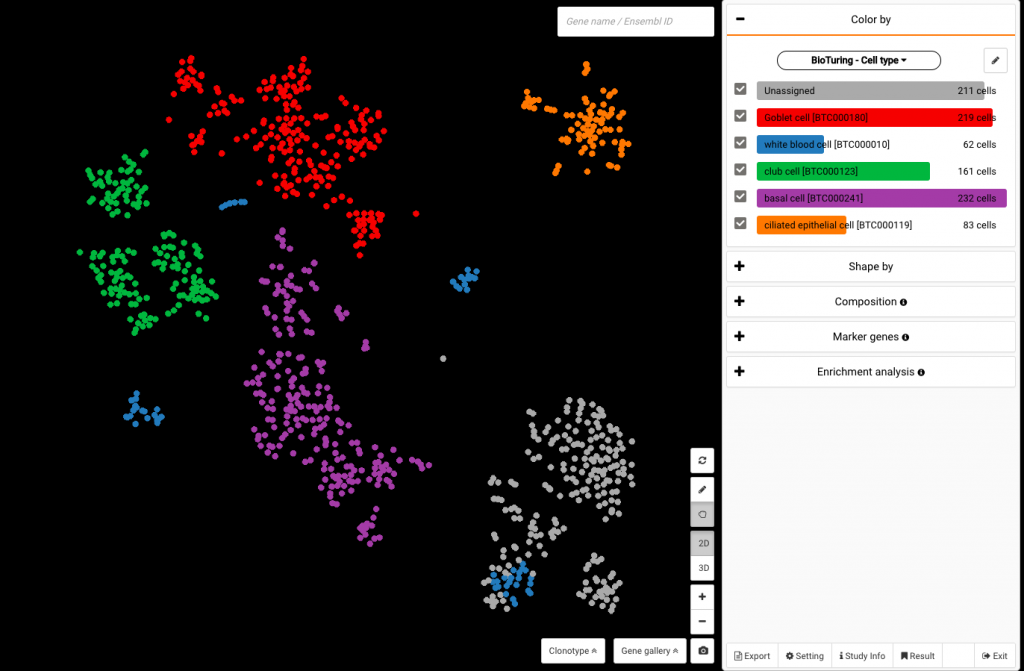
The data set colored by BioTuring’s standardized cell type annotations with ontology numbers
View the expansion of goblet cells and depletion of club cells in smokers’ bronchi
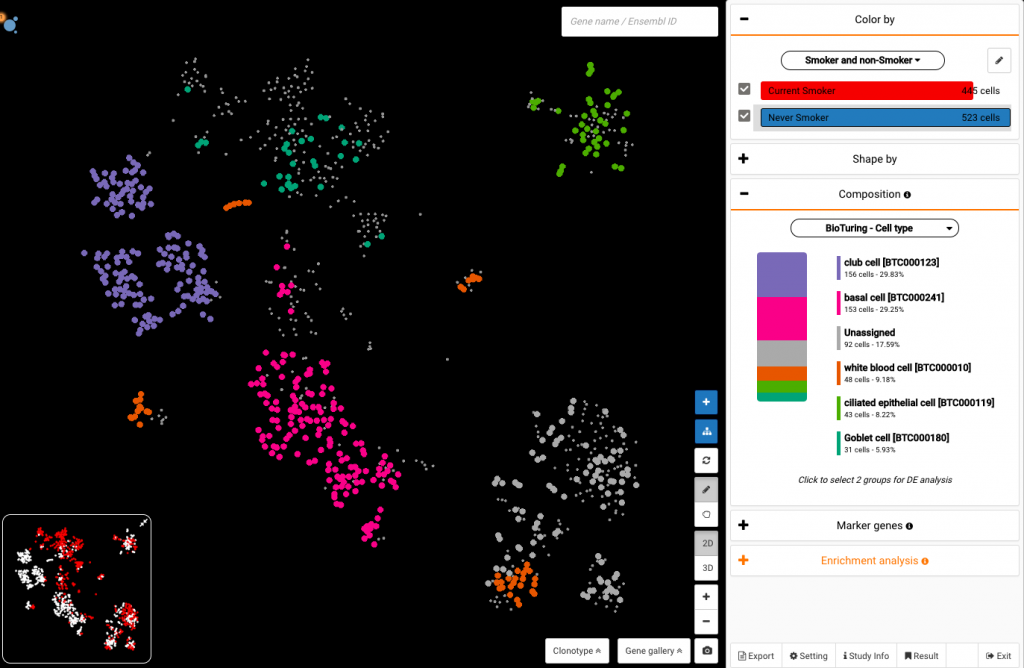
How do cell type composition changes under the effects of smoking? To answer this question, let’s use the Composition box.
First, you need to color your data by Smoker and non-smoker so that you can select them from the Color by panel. After that, choose Never smoker, and expand the Composition section. Click the drop-down under this part and view the composition by Cell type. You should see that never smokers had high percentages of club cells (29.83%), and low shares of goblet cells (5.93%).
Select Current Smoker on the Color by section to view its compositional changes compared with never smokers. Interestingly, the smoking group showed a repolarization in the cell type composition: extremely high percentages of goblet cells, while club cell ratios were low.
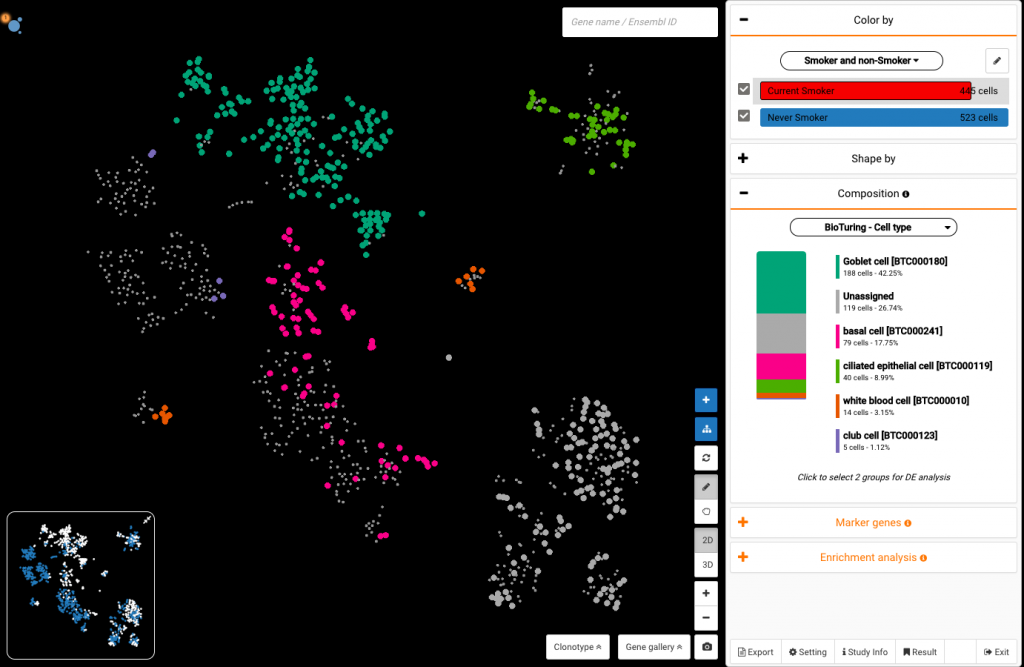
Compositional study of cells from smokers (top) and non-smokers (bottom) shows an expansion of goblet cells and reduction of club cells in smokers
View the smoking-induced detoxification program in smokers’ ciliated cells
To see if the transcriptional profiles of ciliated cell populations changed due to smoke exposure, let’s perform a differential expression analysis of the smokers’ ciliated cells versus non-smokers.
Just color your data by Cell type and select Ciliated cells. The next step is to expand the Composition box to view its composition by Smoker or non-smoker.
Finally, click Run DE analysis. BBrowser will show its differential expression results with an interactive volcano plot, an interactive violin plot, main scatter plot and DE genes’ info plus enrichment analysis results. Among the up-regulated genes are ALDH3A and AKR1C1, encoding aldehyde dehydrogenases and aldo-keto reductases, capable of breaking down tobacco smoke–derived chemical compounds, such as toxic aldehydes and ketones.
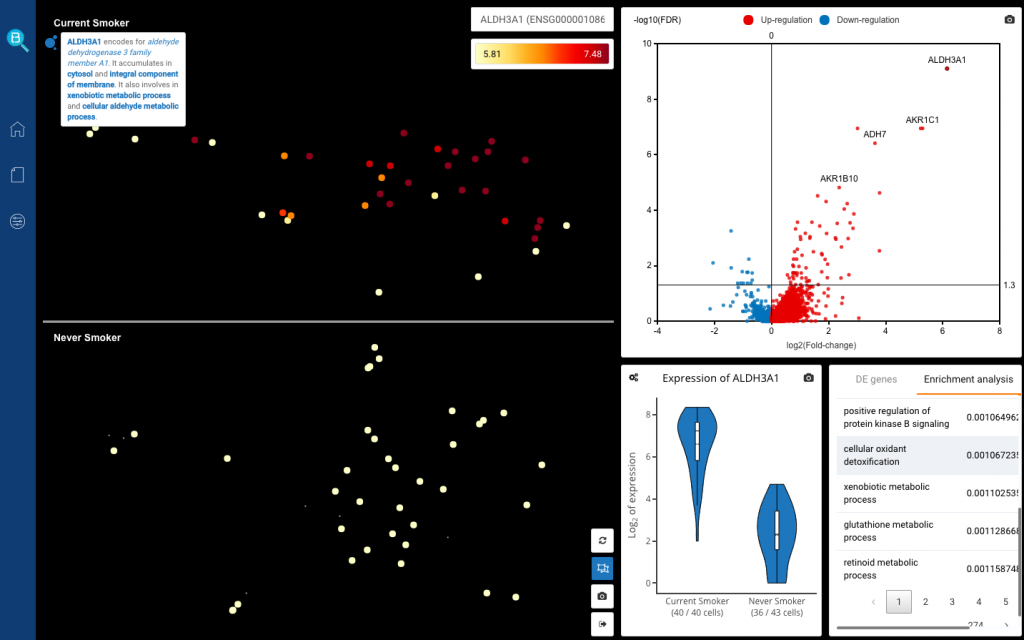
Differential expression analysis of ciliated cells from smokers vs. non-smokers
Click the genes on the volcano plot to show their expression across the 2 groups. Click Split View (at the bottom right corner – the second button) to split the cells from 2 groups of donors so that you can easily observe their gene expression levels.
In addition to the analyses we listed above, you have many more analytical options to explore the dataset in BioTuring Browser, including marker gene finding, enrichment analysis, sub-clustering, querying dual gene expression. Enjoy your time exploring the data!
About BioTuring Cellpedia series:
This is our new blog series that provides you with an overview of some interesting datasets indexed in BioTuring Browser public repository, a platform for instant access and reanalysis of published single-cell RNA-seq data. Details on their experimental designs and key findings are manually curated and presented here, together with brief instructions on how you can explore them in BioTuring Browser.
Hope you will enjoy the series!
Reference:
Duclos, Grant E., et al. "Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution." Science Advances 5.12 (2019)