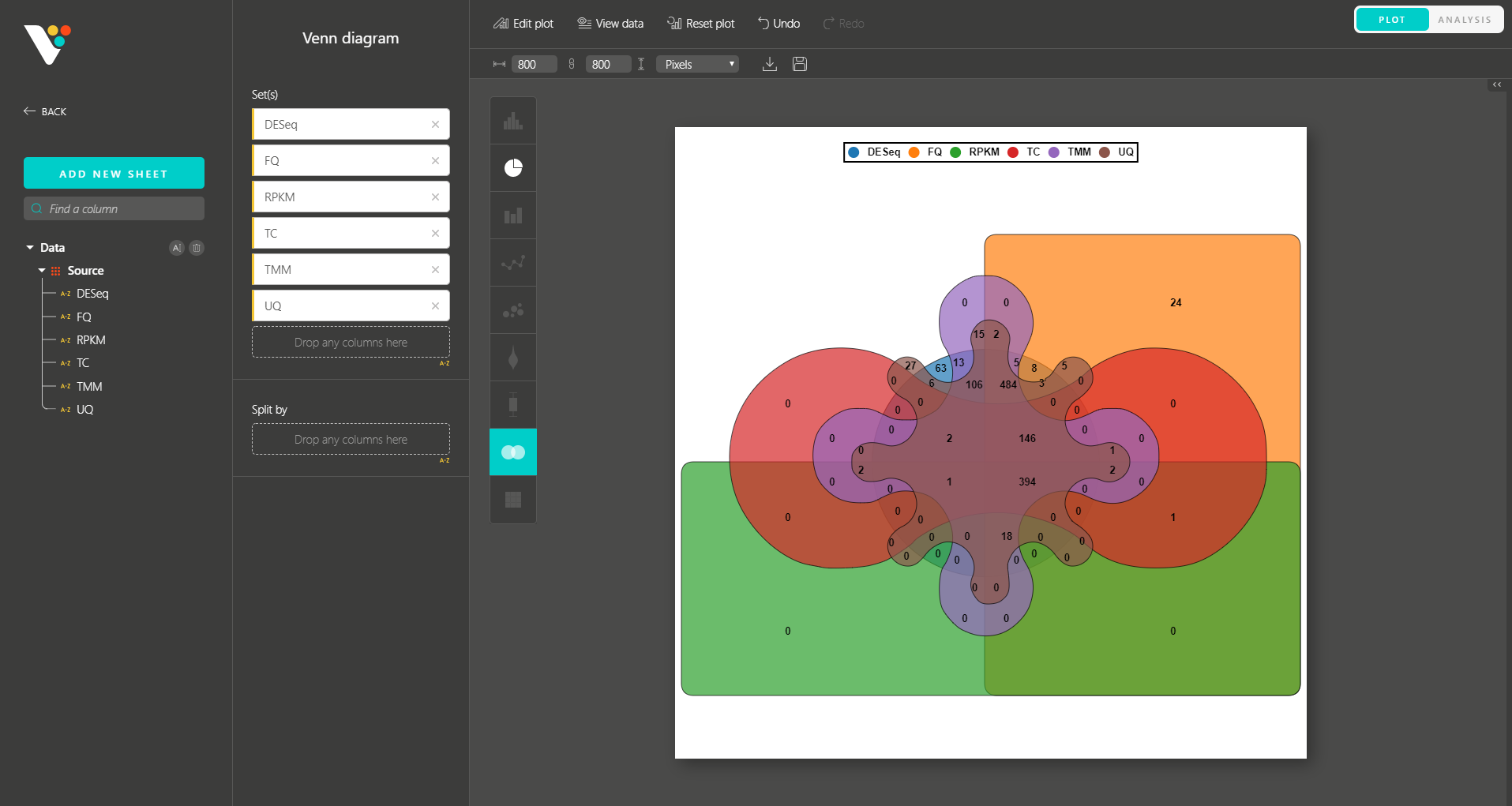
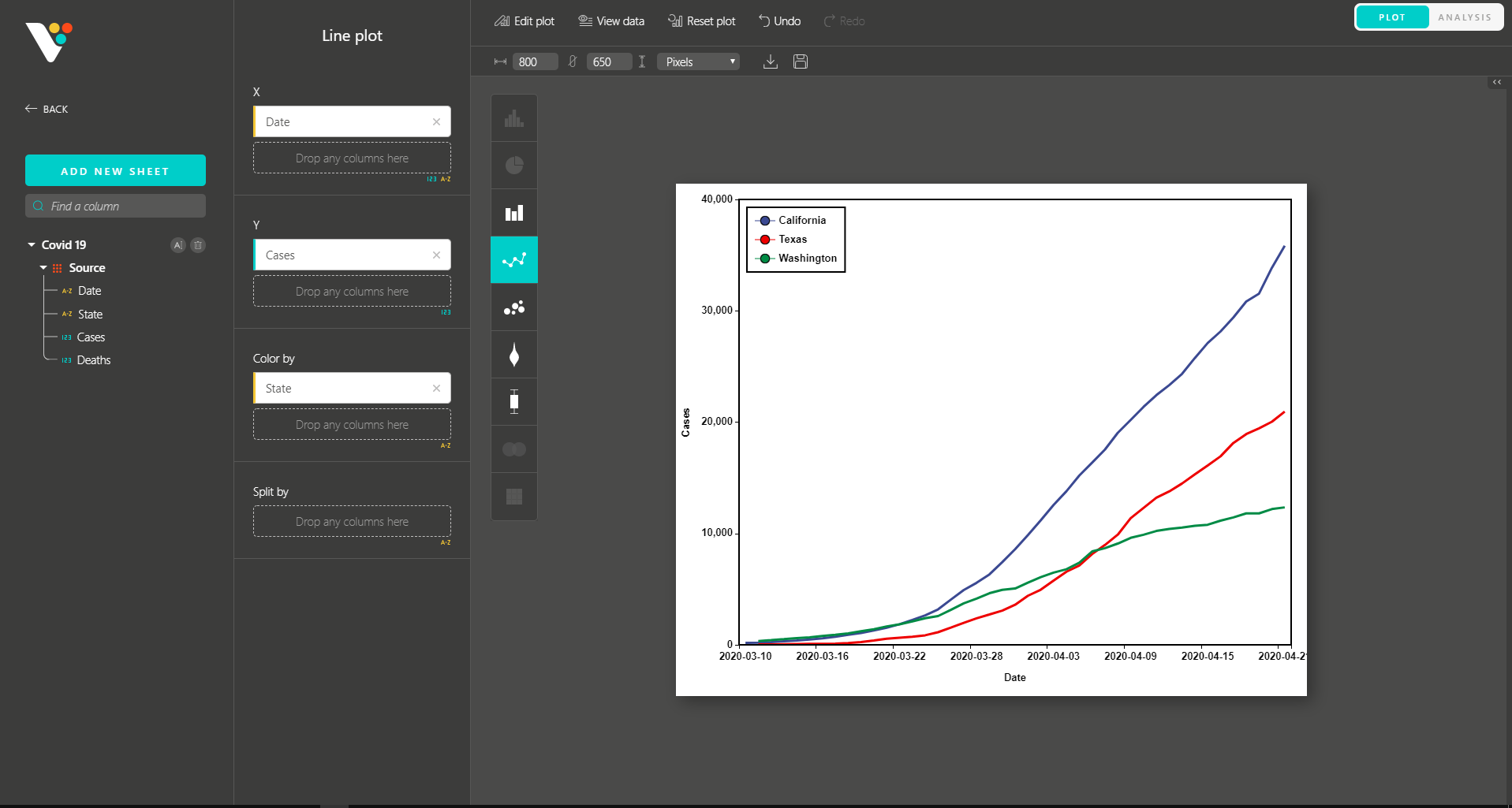
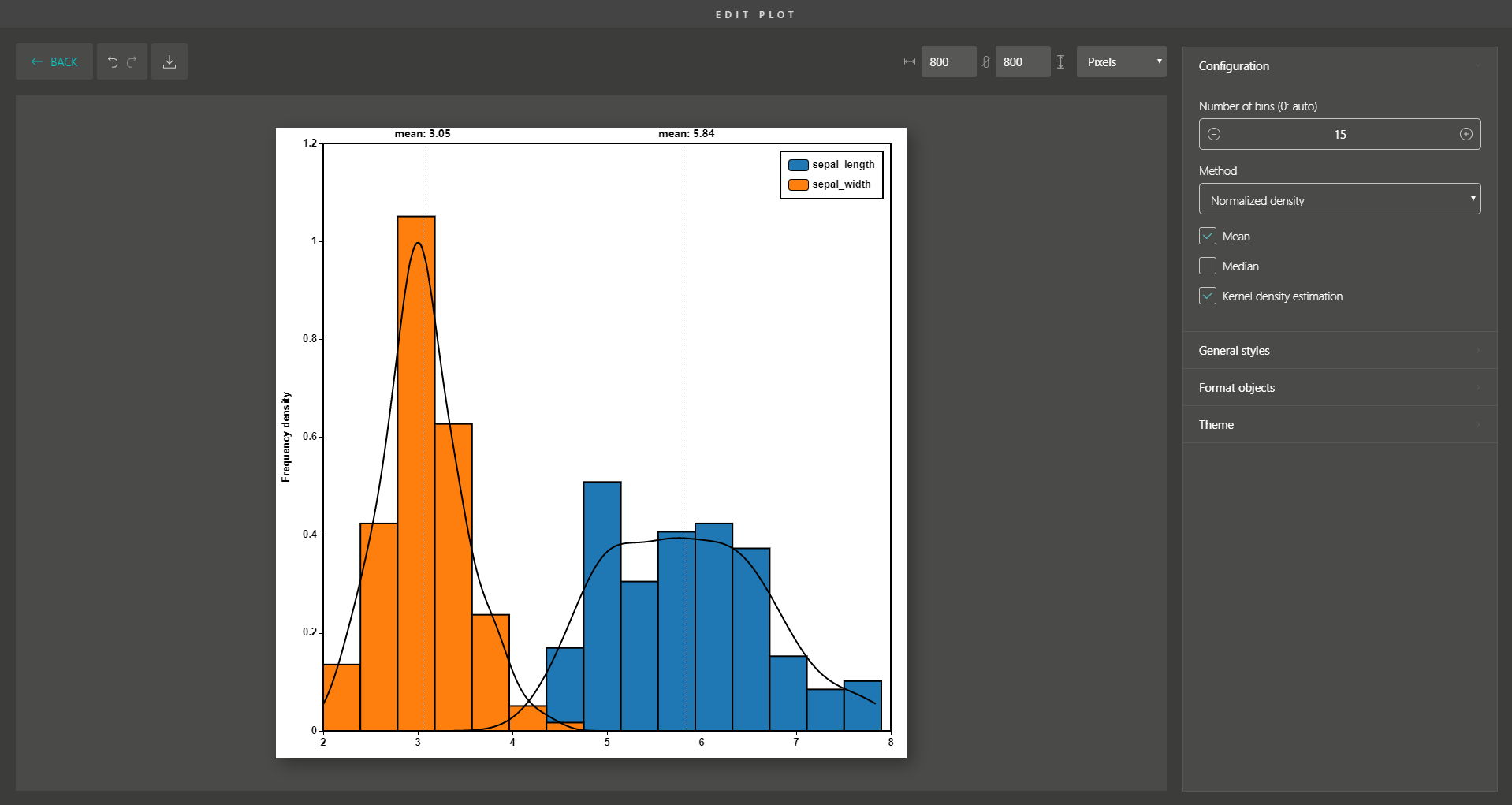
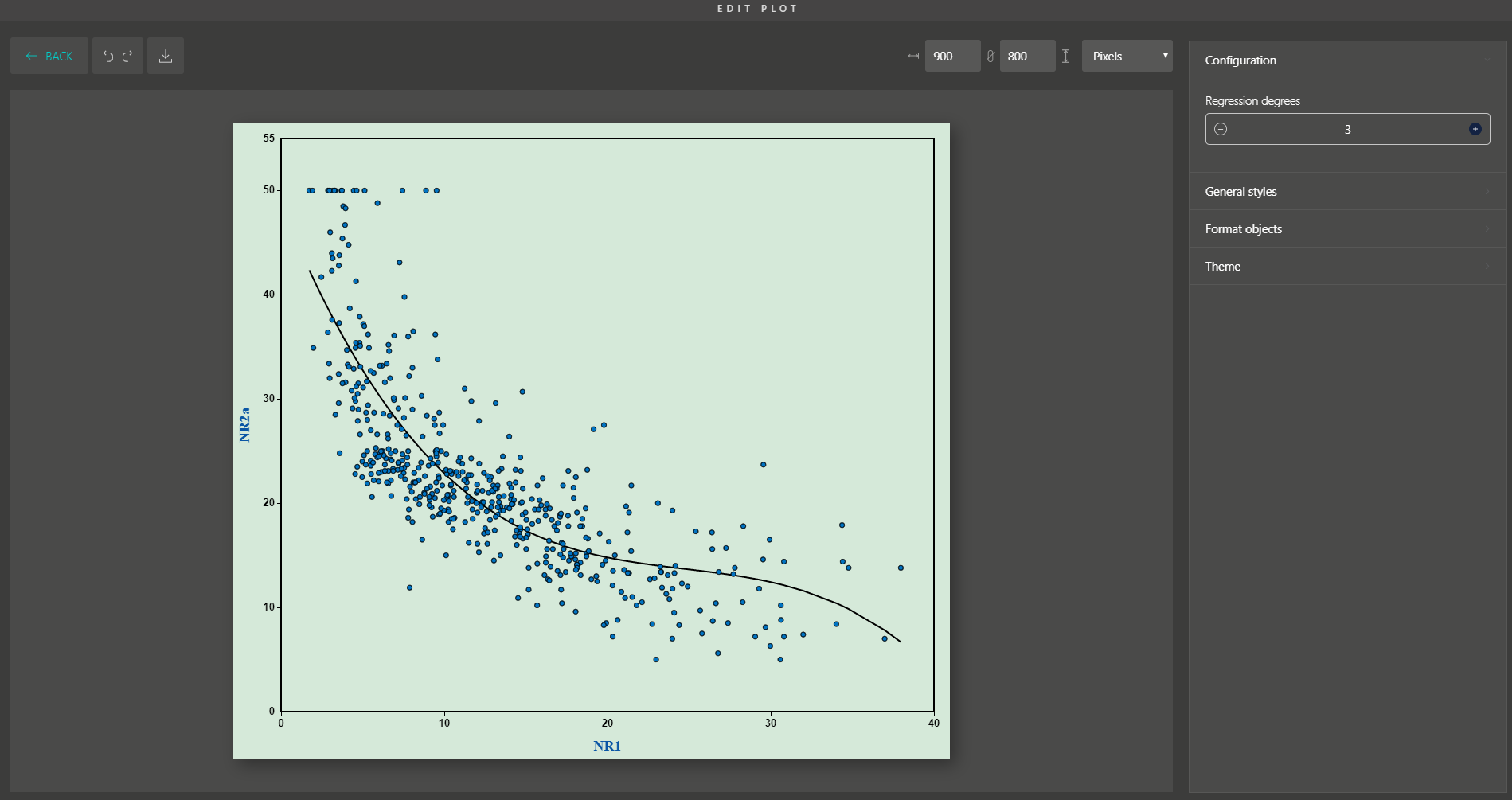
“How to extract the biology behind this huge matrix of thousands of rows and columns?”
Large numbers of data points hold valuable biological stories yet are tricky to graph and analyze. Imagine you had the data for 1000 features and observations, your mind would be full of questions, wondering how to treat it in the right way, what is the go-to visualization method for it, and how to identify the most important features.
To make high-dimensional data visualization and analysis less tricky, we developed BioVinci 2.0, a package that brings automated machine learning and modern visualizations to life sciences. You can easily get BioVinci to recommend the best-fit visualization method for your high-dimensional data, and automatically extract the important features to help you interpret the biological stories.
BioVinci 2.0 also inherits the interactive drag-and-drop interface and diverse plot editing options from Version 1.0 , allowing you to make your biological insights publication-ready.
How can BioVinci 2.0 support you?
Automatically find the best dimensionality reduction method for your data
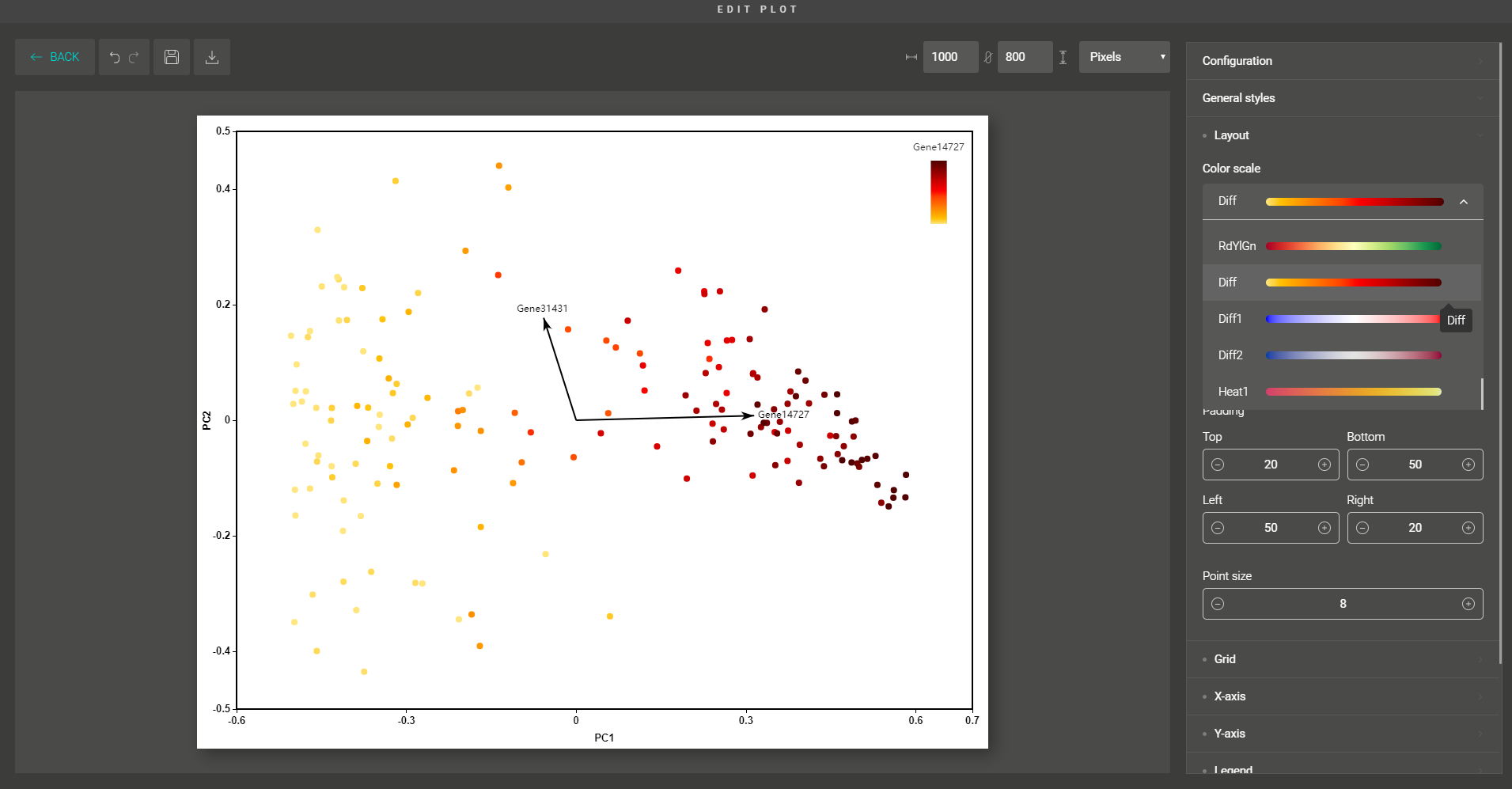
You don’t need to think about which visualization method fits your high-dimensional data best. BioVinci is here to automatically find the best algorithm to convert your thousand-dimension data into 2D and 3D visualizations, such as PCA, UMAP, t-SNE, ISOMAP, and LLE.
PCA-Principal component analysis, a method that transforms data to a new coordinate system by projecting each data point onto a group of orthogonal axes. Read our blog explaining PCA here.
t-SNE – t-distributed Stochastic Neighbor Embedding, a well-suited method for visualizing high-dimensional data in two or three dimensions.
UMAP – Uniform Manifold Approximation and Projection for Dimension Reduction, a method has a better presentation with geographic data and processing speed than t-SNE.
ISOMAP, a method is to compute a quasi-isometric, low-dimensional embedding of a set of high-dimensional data points.
LLE – locally linear embedding, is used to discover nonlinear structure in high-dimensional data by utilizing the local symmetries of linear reorganization.
Extract important features and explain your data using a decision tree model
Upon importing your data for dimensionality reduction, BioVinci will automatically detect the most important features and explain your data using a decision tree model. The function helps you to easily spot the key features (eg. genes, proteins, peptides,…) that classify your data.
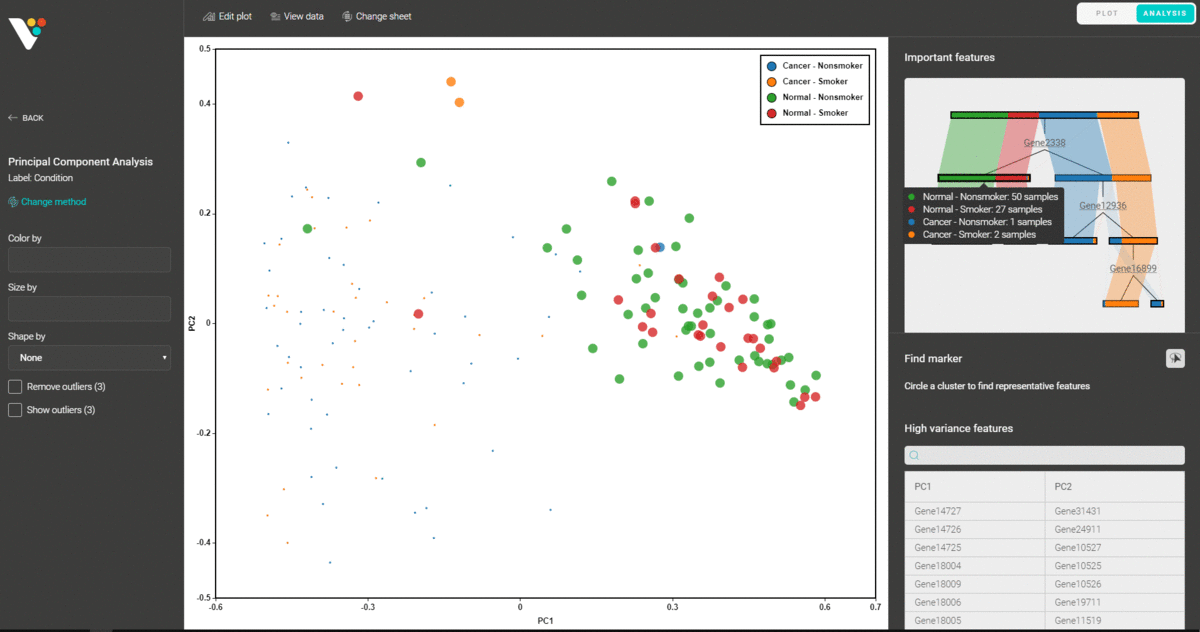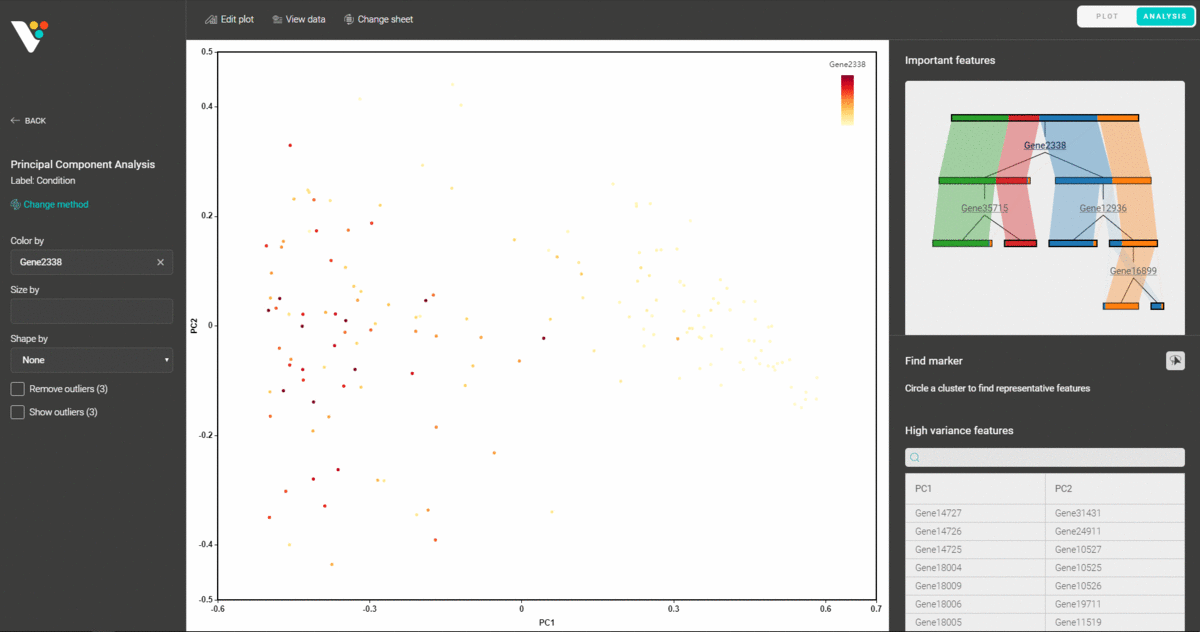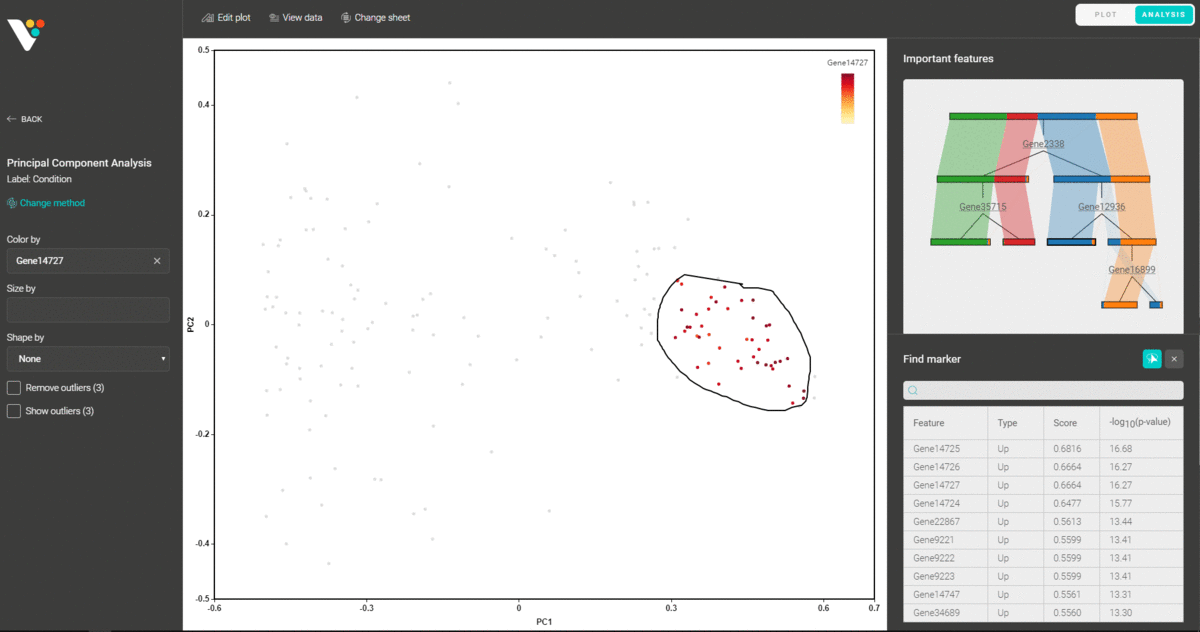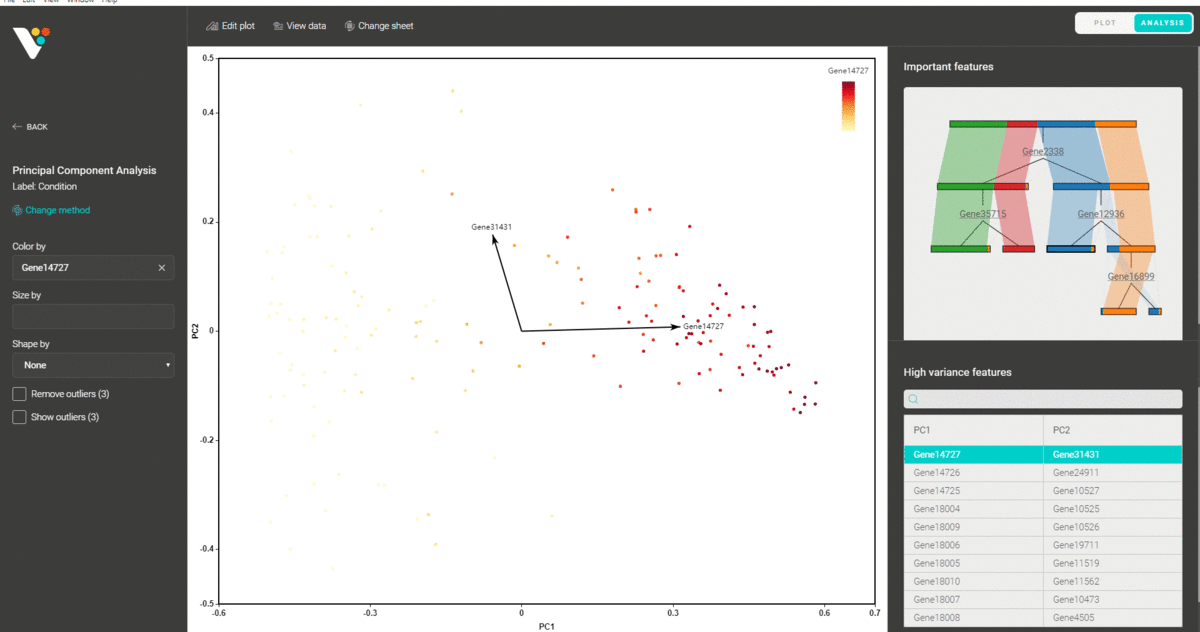
You can also circle any group of data points and find the representative features that distinguish them from the remaining groups.
Easily customize output plots for publication and enjoy other modern plot options
Besides, you are supported with a variety of plot editing options to turn your dimensionality reduction results into high-quality figures for publication. From coloring, sizing, to customizing the legends, every action can be easily done by simple mouse clicks.
Similar to Version 1.0, you can simply drag and drop to create other modern visualizations of your data. BioVinci is now available for download at https://vinci.bioturing.com
–
Take a look BioVinci citations [here]