Introduction
Why are tumors so resilient against available cancer therapies? One answer lies in intratumoral heterogeneity.
Intratumoral heterogeneity describes the diversity in tumor cell populations. Cancer cells exhibit startling distinctions in their types, shape, metabolic activities, transcriptomic profiles, and more. The causes of intratumoral heterogeneity are both heritable (clonal expansion and tumor evolution) and non-heritable (cancer stem cells, phenotypic elasticity) [6].
The existence of intratumoral heterogeneity implies that there could be no one-size-fits-all solution for cancer treatments, for tumors possess incredible elasticity and adaptability. It’s time for researchers to look at cancer cells at a different angle–not as a block of tissue, but as single cells with individual characteristics. The demand for such a close-up into tumor heterogeneity makes single-cell RNA-sequencing (scRNA-seq) a powerful tool for oncologists. Indeed, there has been a growing number of studies dissecting intratumoral heterogeneity by various scRNA-seq platforms.
From the BioTuring public database, we picked out 5 interesting scRNA-seq datasets about liver cancer–one of the most common malignancies worldwide (Aizarani et al., 2019, Sharma et al., 2020, Zhang et al., 2019, Zheng et al., 2017, Ma et al., 2019). These datasets examined liver cancer samples from a diverse collection of live cancer types, patient status, histology, etc. and compared them to a comparably diverse collection of normal tissues, unveiling unique features rooted in intratumoral heterogeneity. What lies in these features are the potential targets that may warrant more effective liver cancer therapies.
With the comprehensive toolbox for scRNA-seq data analysis from BBrowser, let’s see what over 200,000 cells in these studies have in common!
Insights
#1 Intratumoral Heterogeneity Present Across Datasets
A highly heterogeneous cell composition is consistently profiled in the three studies of Aizarani et al. (2019), Sharma et al. (2020), and Ma et al. (2019) (Figure 1). While each study captured the landscape of liver cancer cell populations at a different angle, the liver cancer-associated cells found can be simply classified into two categories: liver-specific and immune cells. For the former, endothelial and hepatocytes are the most commonly found groups. Others include Kupffer cells, fibroblasts, erythroids, and more specific endothelial cell subtypes.
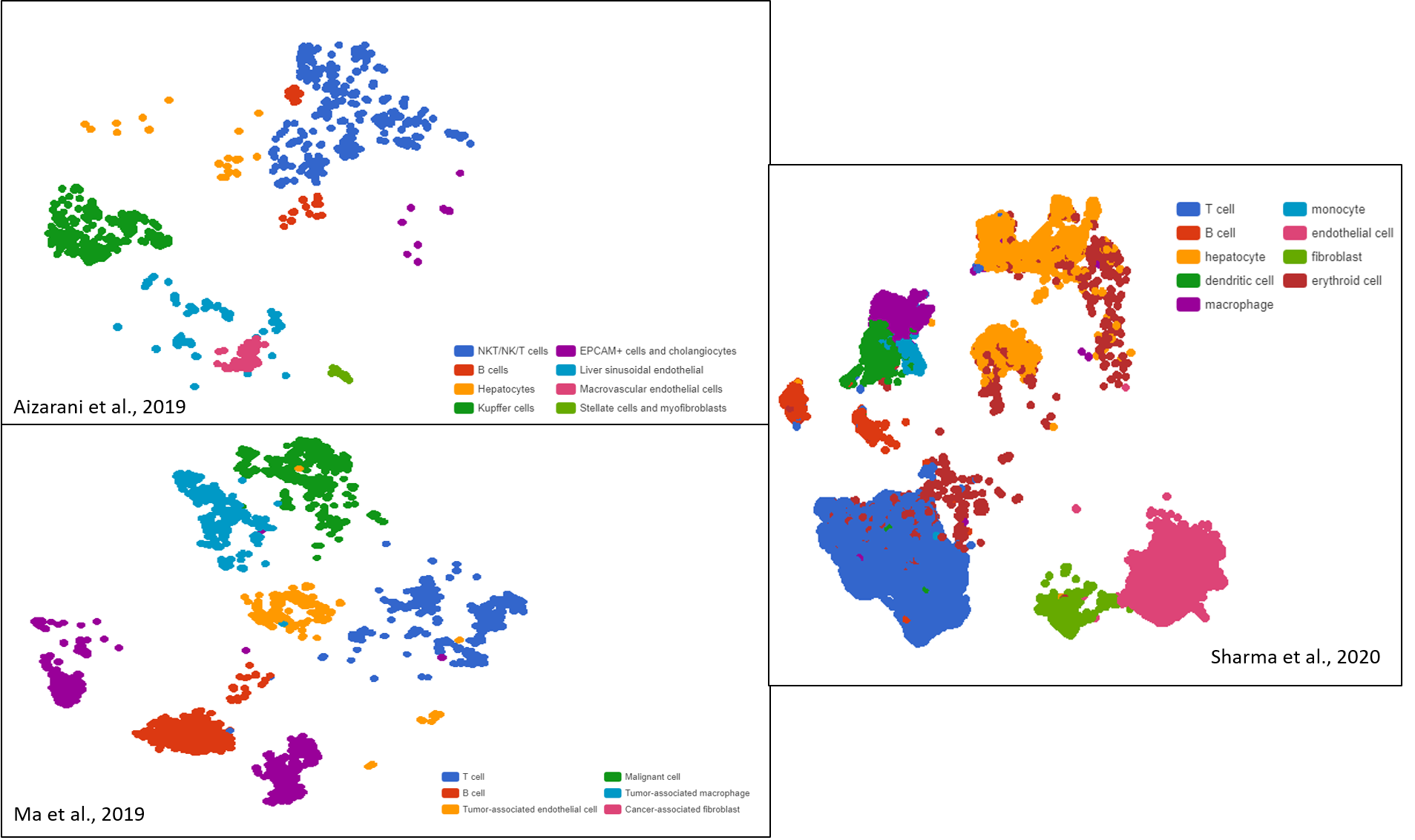
Figure 1. Liver cancer cell compositions of three different datasets exhibit high diversity, visualized as t-SNE and UMAP plots by BBrowser
For the latter category, an interesting common point emerges when putting all datasets together: immune cells always take a significant portion (30-50%) in tumor cell composition (Figure 2). These immune cells are of both innate and adaptive origins. The majority of innate immune cells present across datasets include macrophages and natural killers, while those of the adaptive immune system are dominated by T cells.
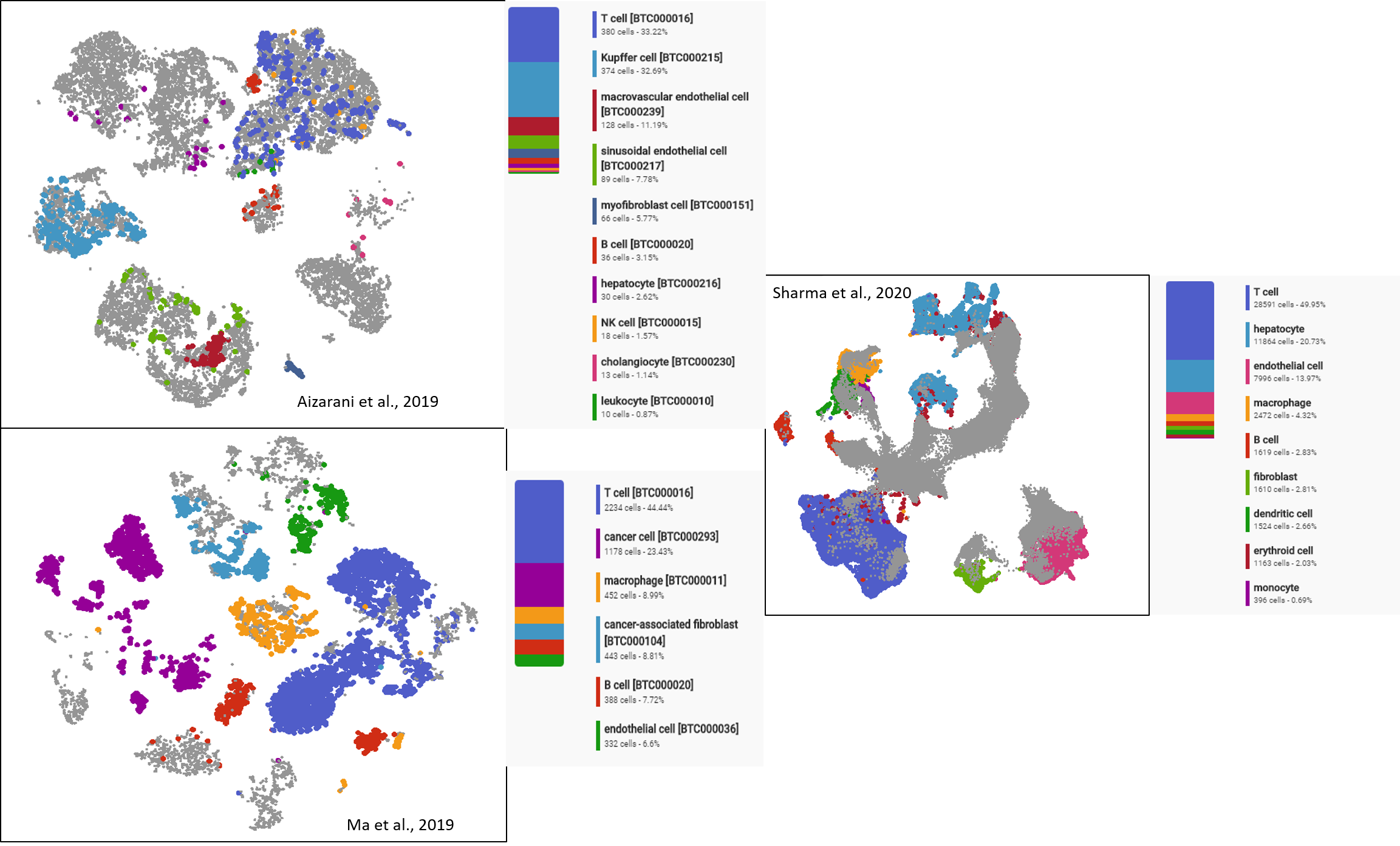
Figure 2. The shared dominance of Immune cells (in which T cells take the majority) in all liver tumors profiled in three different datasets.
In addition, these studies suggest the enrichment of immune cells in tumors compared to normal samples, and this subset exhibits a note-worthy level of intratumoral heterogeneity as well. That leads us to insight #2…
#2. Dynamic Changes in The Immune Cell Subset of Liver Tumors: From Composition to Transcriptomic Profiles
Zhang et al. (2019) and Zheng et al. (2017) took a deeper look into the immune cell subset of liver cancer samples and unveiled another layer of intratumoral heterogeneity in these populations. There are several changes in the composition and gene expression of immune cells between normal and malignant states, probably reflecting the busy immune processes inside liver tumors [2,3,4].
In particular:
-
Suppressed and exhausted T cells found in liver cancer: tumors enrich CD4+regulatory T and exhausted-type CD8+ cytotoxic T cells
Being the dominant cell type in the immune cell subset, the T cell subset attracted most attention. Three studies suggest a clear enrichment of CD4+ regulatory T (Treg) cells and the decline or exhaustion of CD8+ cytotoxic T cells in tumors (Figure 3) [2,3,4].
These results suggest a stronger immunosuppressive state within liver cancer, both by increasing Treg activities and decreasing cytotoxic T cells’ surveillance. 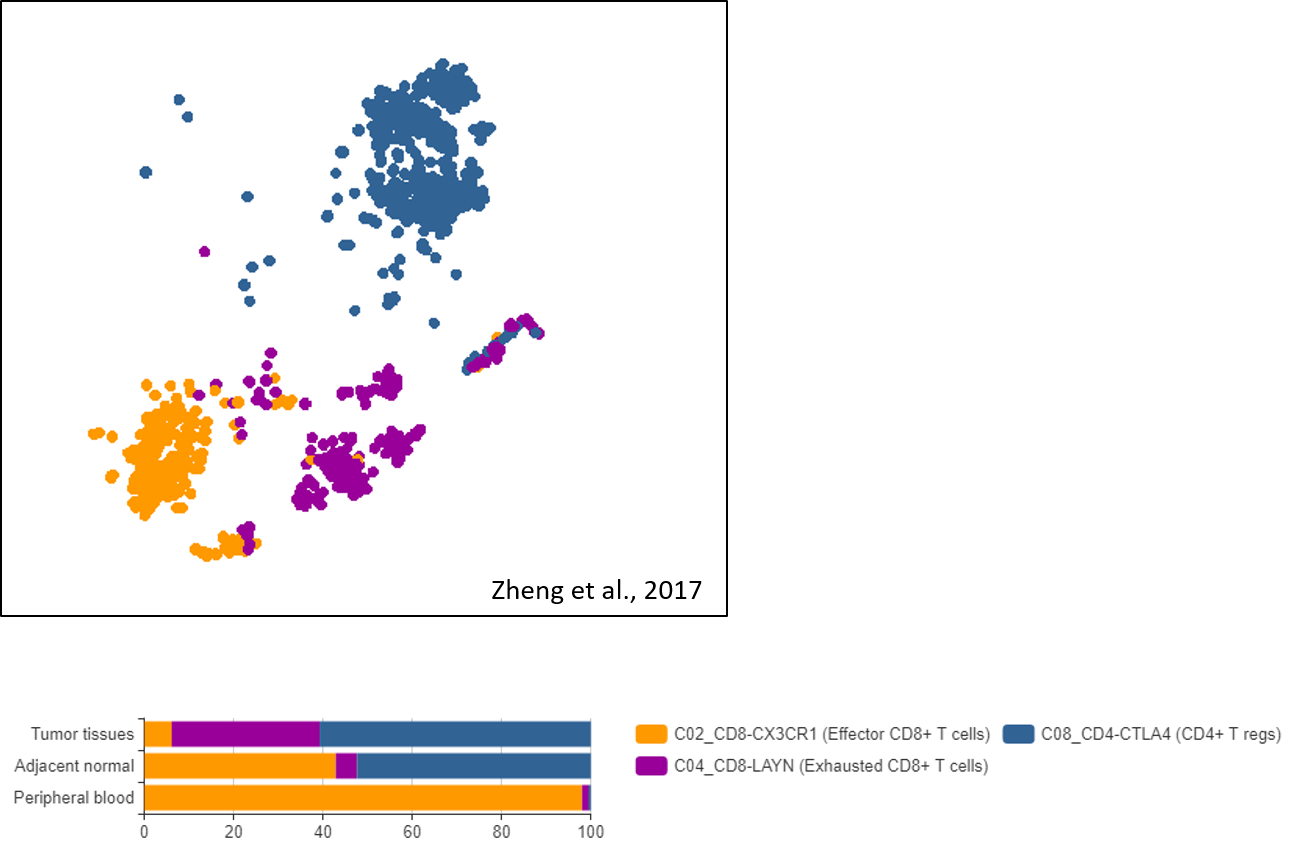 Figure 3. Important T cell subsets and their enrichment in liver tumors found in the dataset of Zheng et al., 2017, visualized by BBrowser
Figure 3. Important T cell subsets and their enrichment in liver tumors found in the dataset of Zheng et al., 2017, visualized by BBrowser
-
The Two Faces of Tumor-enriched Macrophages Seen Through Different Transcriptomic Profiles
Among six macrophage clusters identified, Zhang et al. (2019) focused on two clusters that are enriched in liver cancer tissue, namely myeloid-derived suppressor-like (MDSC-like) and tumor-associated macrophage-like (TAM-like) (Figure 4). Differential gene expression analysis draws a clear border between the transcriptomic profiles of the two clusters, implying that liver tumor-enriched macrophages exist in two states.
What those different states mean is still an open question, but the authors are much interested in the latter cluster, which resembles TAM-like macrophages reported in other cancers. More excitingly, their transcriptomic profiles contain gene signatures that highly correlates with poor patients’ survival and were profiled in lung cancer, namely APOE, C1QA, C1QB, TREM2, SLC40A1, etc. It’s potential that this TAM-like cluster will be of high therapeutic value in multiple malignancies once it’s better characterized.
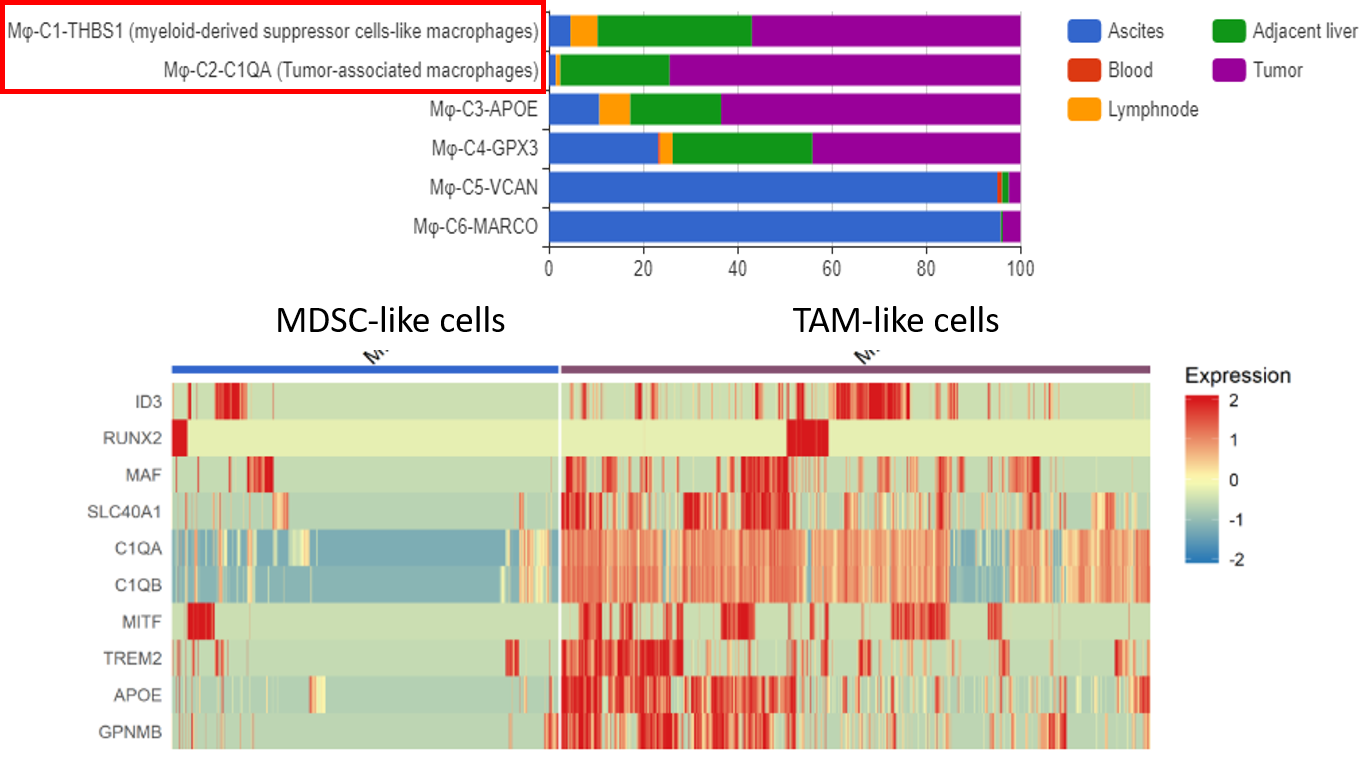
Figure 4 The two macrophage subtypes enriched in Tumors (upper panel) and their distinct expression profiles (lower panel). Data provided by Zhang et al., 2019, cell composition and gene expression analysis done by BBrowser
-
A never-known-before Dendritic cells subset enriched in liver cancer
In the same study, Zhang et al. (2019) profiled four dendritic cell (DCs) clusters, one of which highly expresses LAMP3+ (DC-c4-LAMP3). What’s unique about these LAMP3+ DCs is that they don’t correspond to previously known DC subsets.
Similar to the TAM-like macrophage cluster, this LAMP3+ DC subset may come out as a valuable therapeutic target, since LAMP3 (and other signature genes) has been found to be highly expressed in several subsets of DCs in breast cancer and lung cancer.
#3 Novel Features of The Tumor Microenvironment Unveiled Through Its Diversity
Sharma et al. (2020) found a striking similarity between some subsets of endothelial cells and macrophages between tumor and fetal liver, namely PLVAP+/NRP1+/VEGFR2+ endothelial cells and FOLR2+ tumor-associated macrophages. This similarity implies that liver cancer may utilize similar fetal processes, such as immunosuppressive and the Notch/VEGF signaling in vascular development [2].
Another interesting insight was provided by Ma et al. (2019), in which the intratumoral diversity exhibited prognosis value [5]. The authors first measured intratumoral diversity through principal components of the transcriptomic profiles, based on which they divided malignant cells into two groups: one highly diverse (Div-High) and one less diverse (Div-Low). Statistical analysis between these two groups and the patients’ conditions suggests a strong correlation between tumors of high intratumoral diversity and a worse overall outcome. This opens an opportunity for scRNA-seq to be a reliable, comprehensive prognosis tool.
Closing
With scRNA-seq, researchers can now access a trove of previously hidden data. They showed us that intratumoral heterogeneity is much more than just diverse cell composition. This diversity encompasses complex tumor features, critical immune processes, and potential therapeutic targets.
However, as the amount of scRNA-seq studies increases, connecting the dots between datasets will be a new challenge. As we have seen, no single study can cover all the layers of intratumoral heterogeneity.
BBrowser and the BioTuring public database are here to help! Our public database now contains over hundreds of scRNA-seq datasets, covering a wide range of immuno-oncology studies. The comprehensive toolbox of BBrowser allows hassle-free data analysis to reproduce published results or to create your own insights. Are you ready to make new discoveries with us?
Want to know how to reproduce these insights? Watch our tutorial for the dataset of Sharma et al. (2020) below
References
- Aizarani, N., Saviano, A., Mailly, L., Durand, S., Herman, J. S., Pessaux, P., … & Grün, D. (2019). A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature, 572(7768), 199-204.
- Sharma, A., Seow, J. J. W., Dutertre, C. A., Pai, R., Blériot, C., Mishra, A., … & DasGupta, R. (2020). Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell, 183(2), 377-394
- Zhang, Q., He, Y., Luo, N., Patel, S. J., Han, Y., Gao, R., … & Zhang, Z. (2019). Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell, 179(4), 829-845.
- Zheng, C., Zheng, L., Yoo, J. K., Guo, H., Zhang, Y., Guo, X., … & Zhang, Z. (2017). Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell, 169(7), 1342-1356.
- Ma, L., Hernandez, M. O., Zhao, Y., Mehta, M., Tran, B., Kelly, M., … & Wang, X. W. (2019). Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer cell, 36(4), 418-430
- Marusyk, A., & Polyak, K. (2010). Tumor heterogeneity: causes and consequences. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 1805(1), 105-117.