The Art of Setting Single-cell Quality Control Parameters
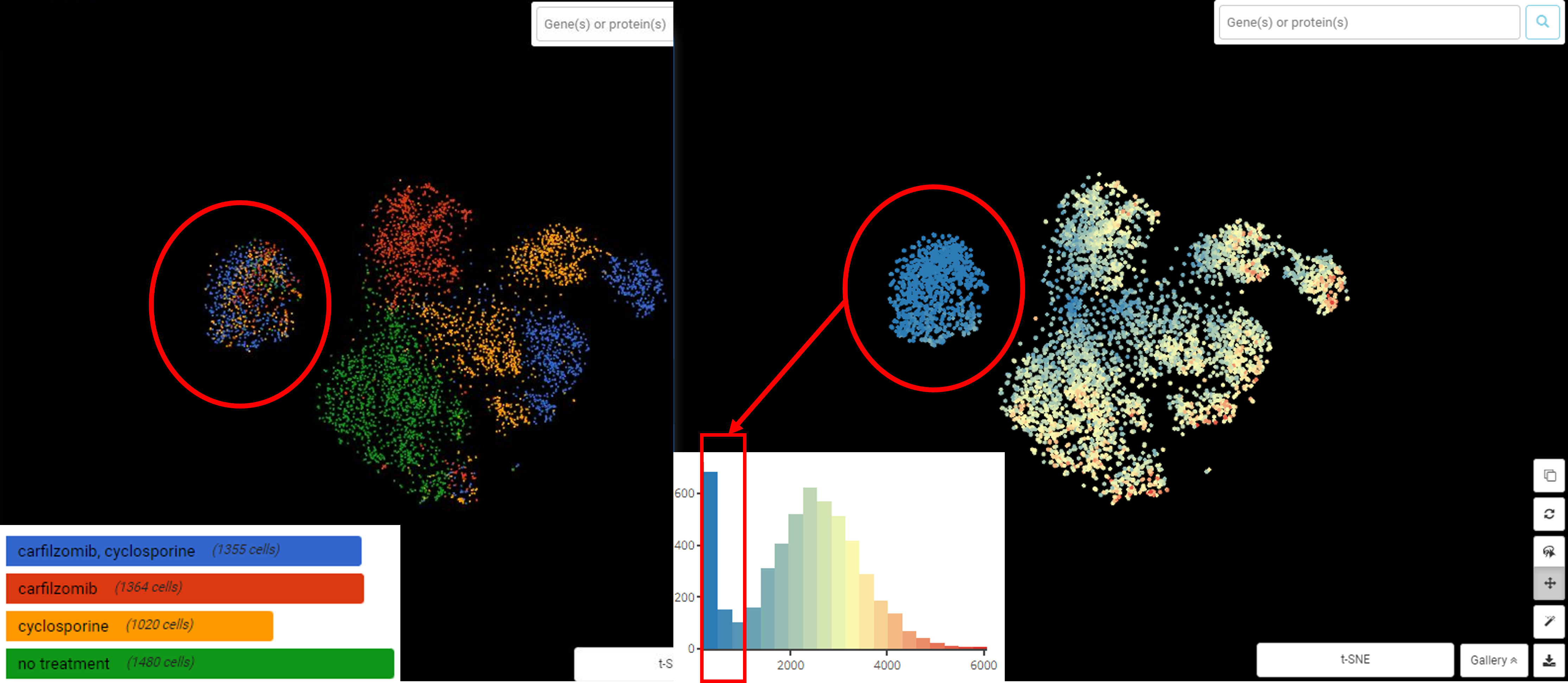
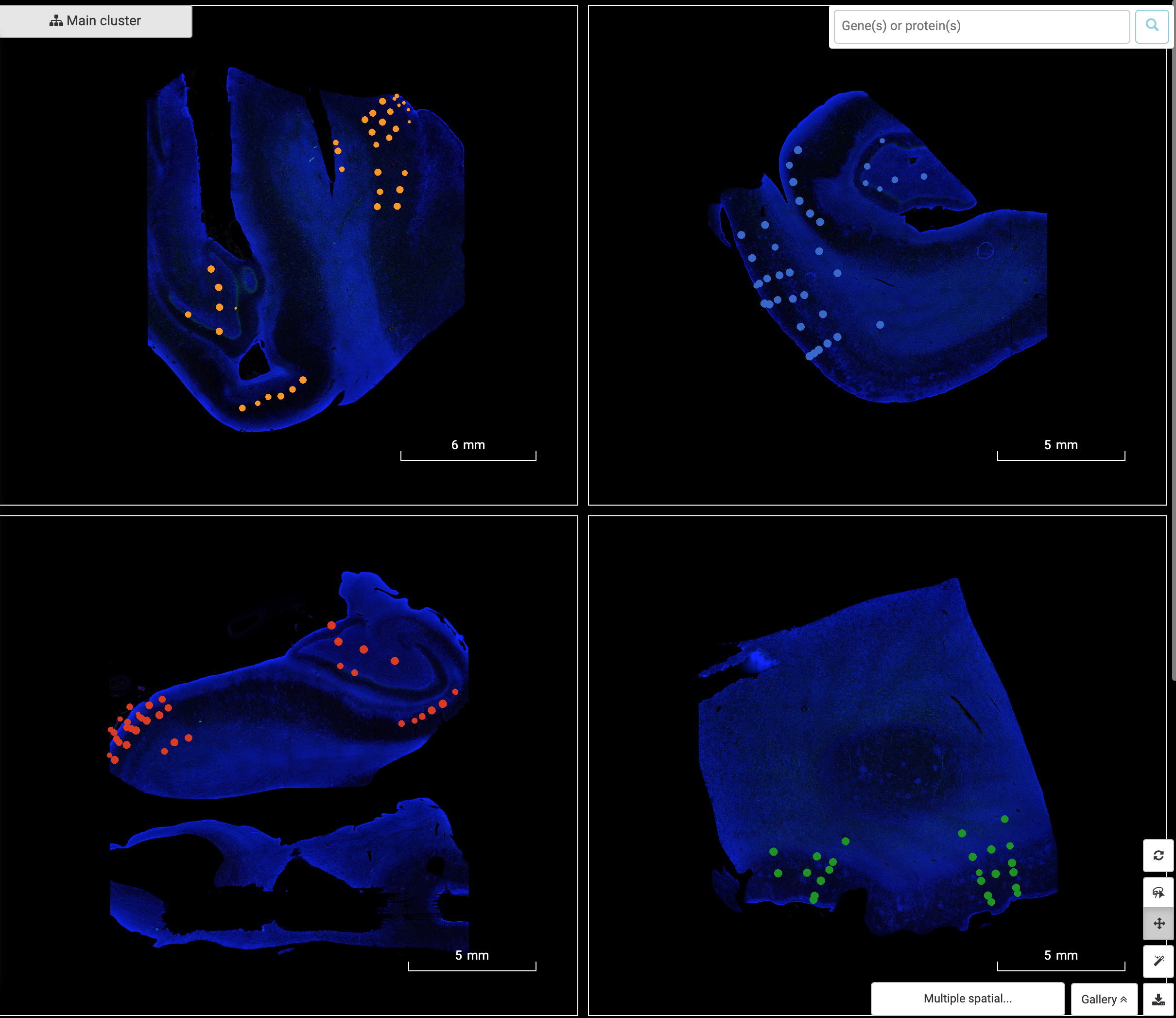
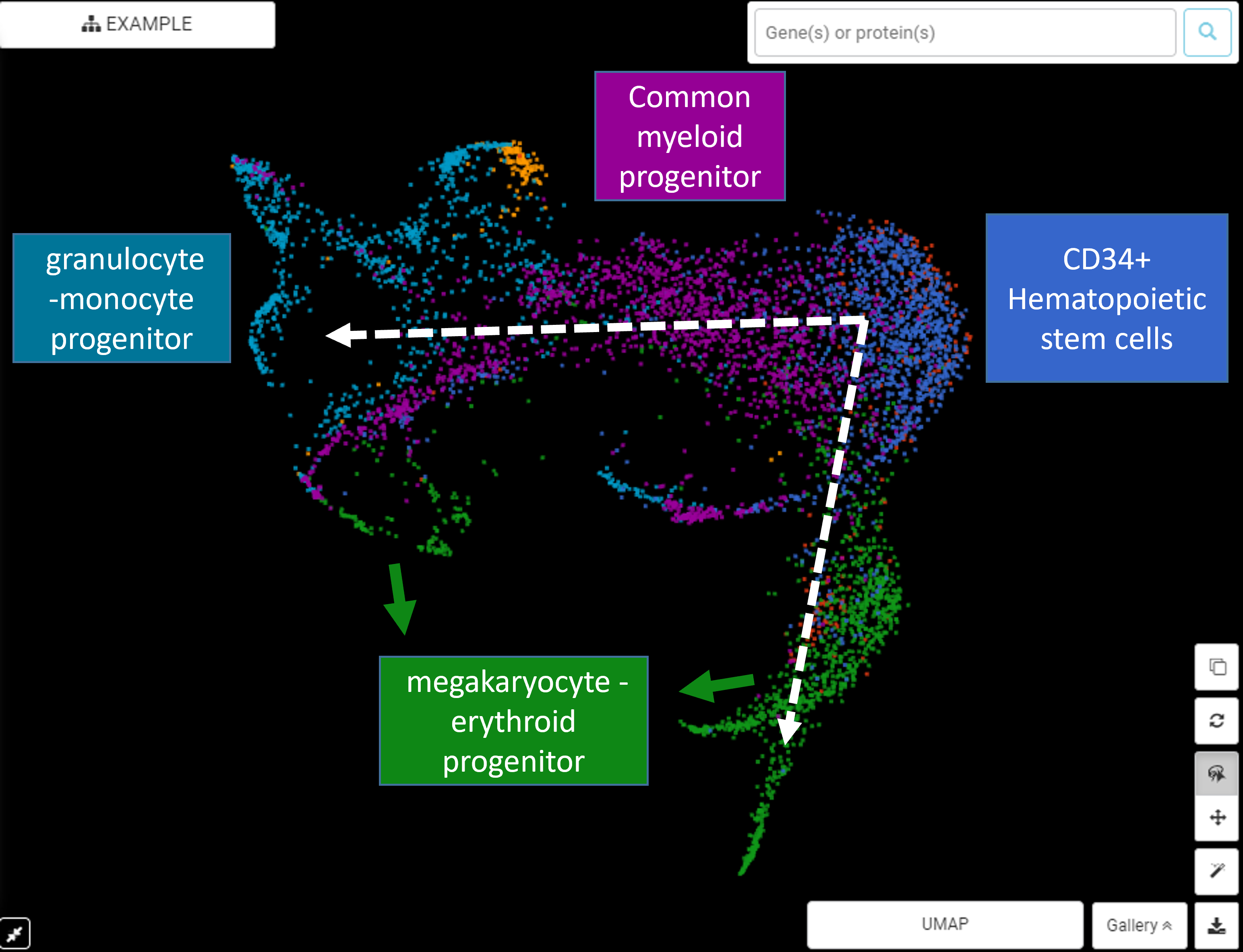
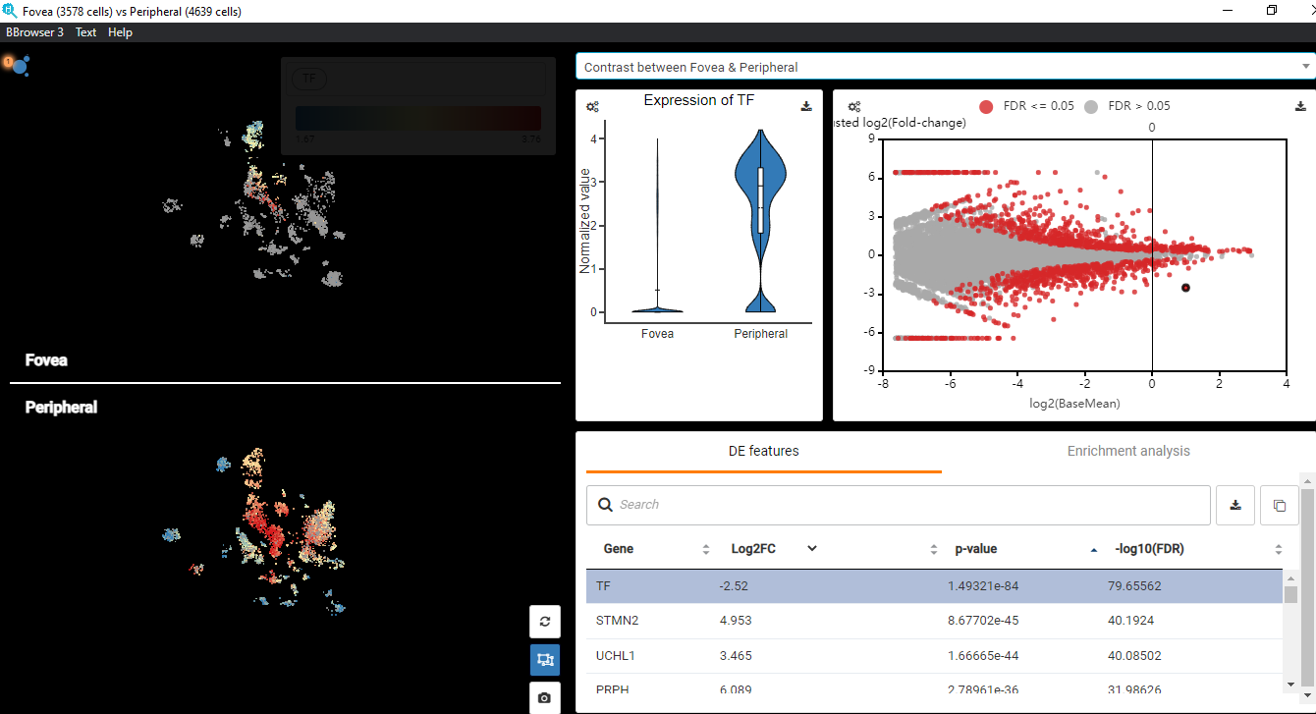
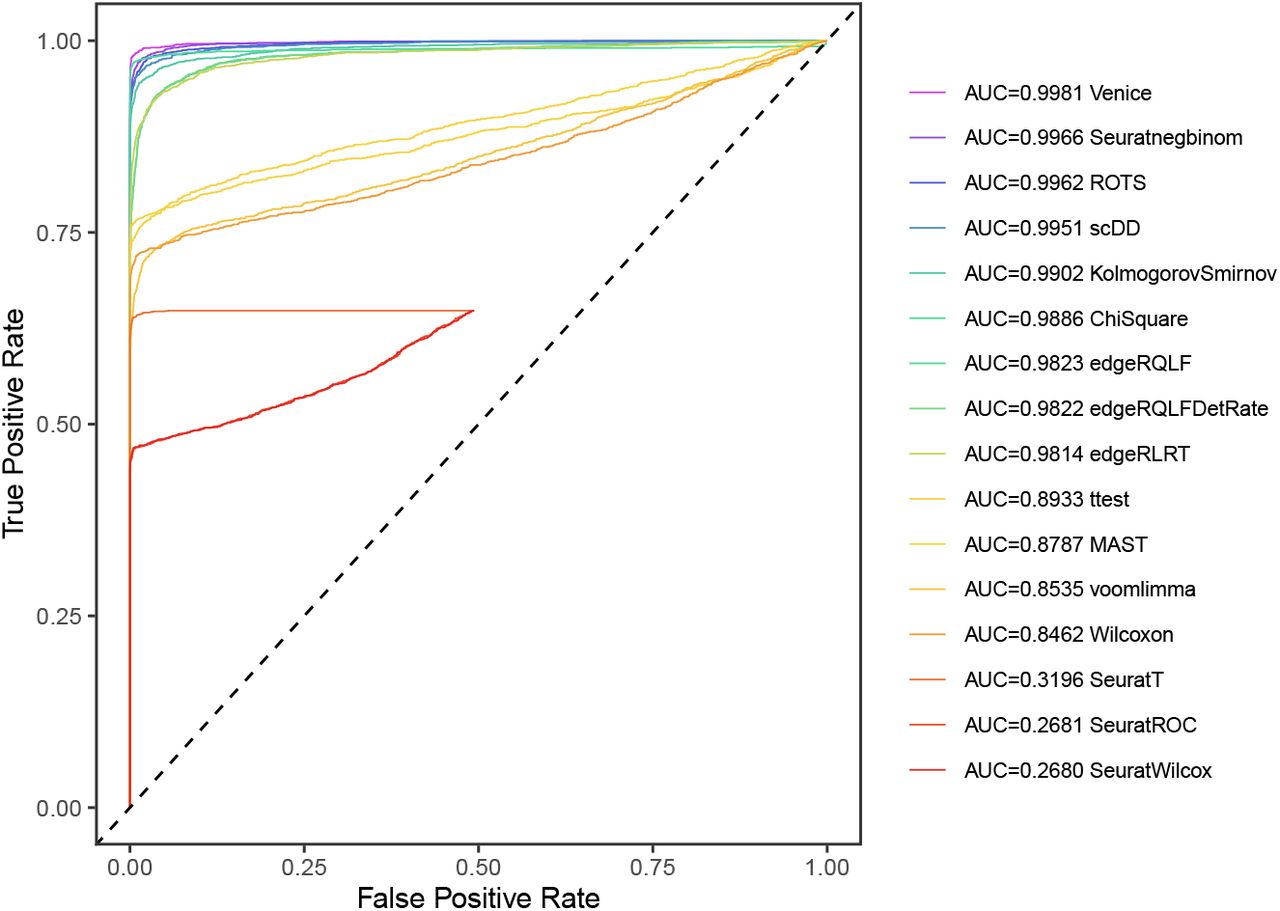
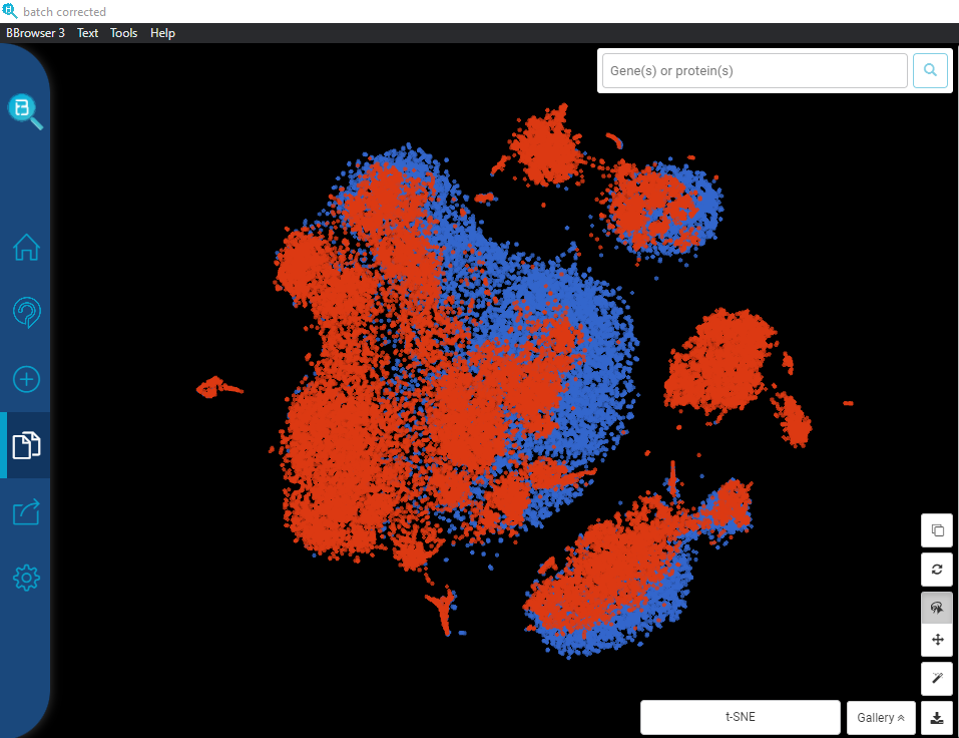
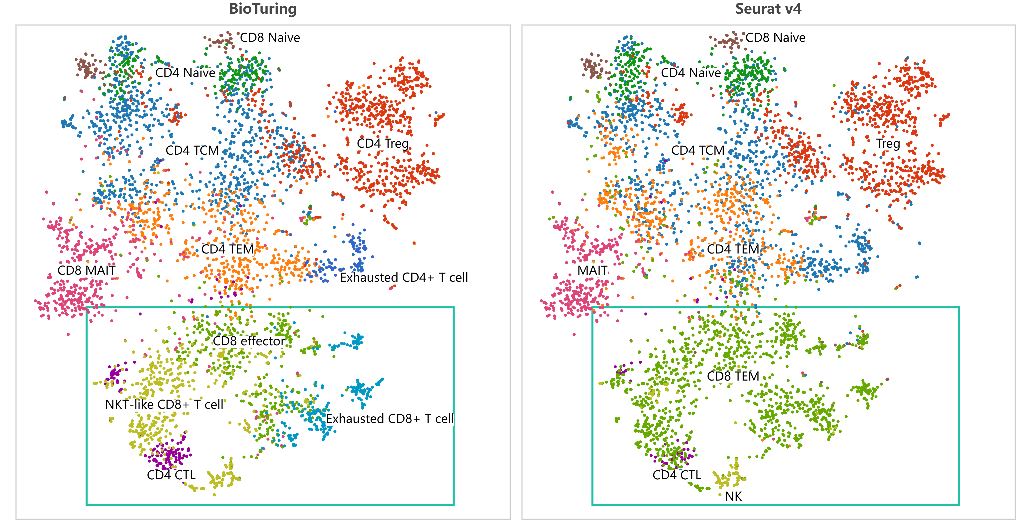
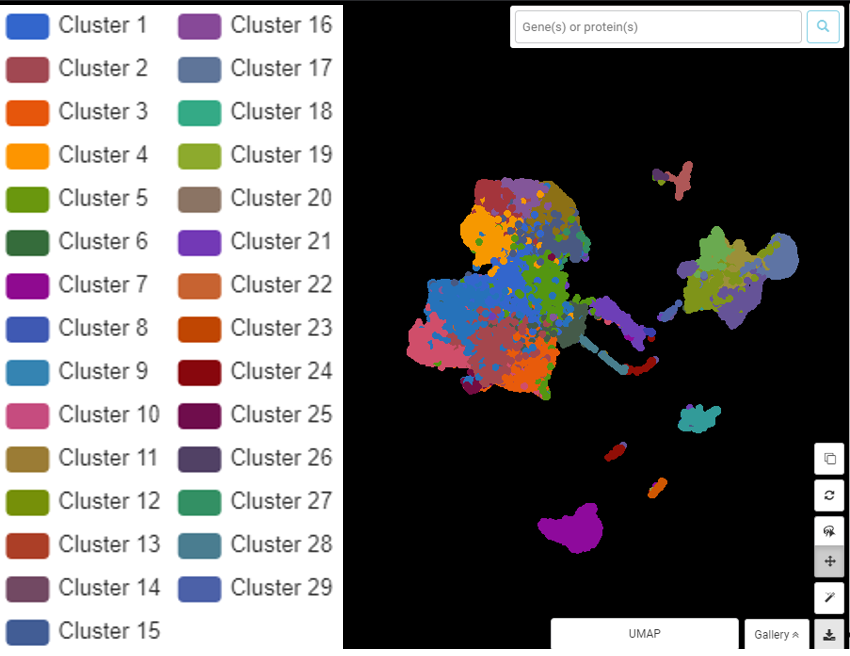
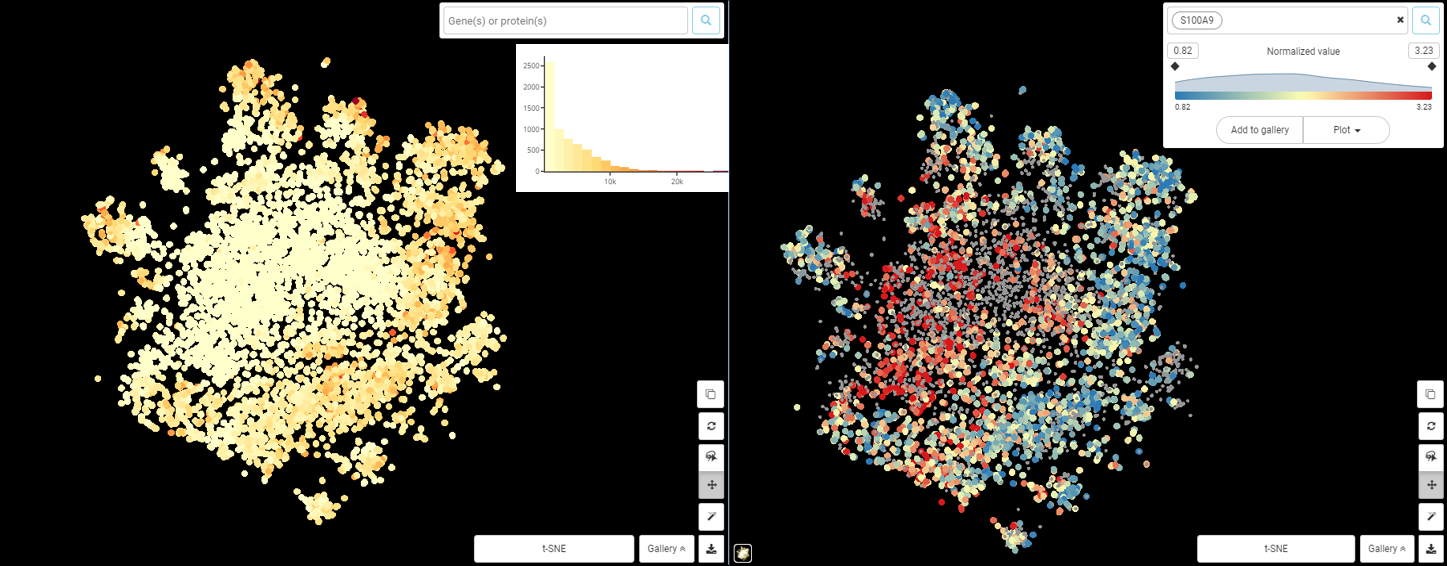
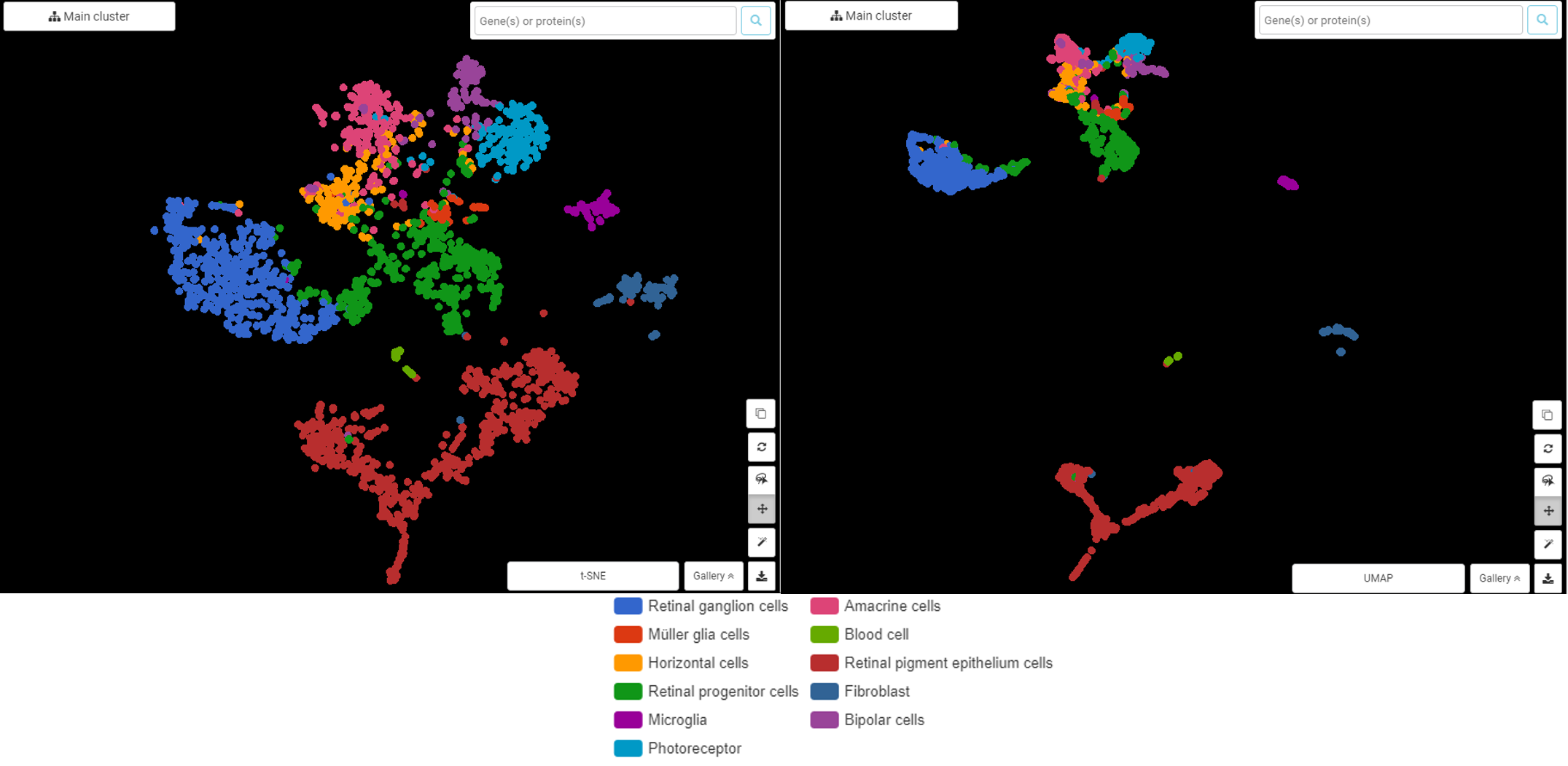
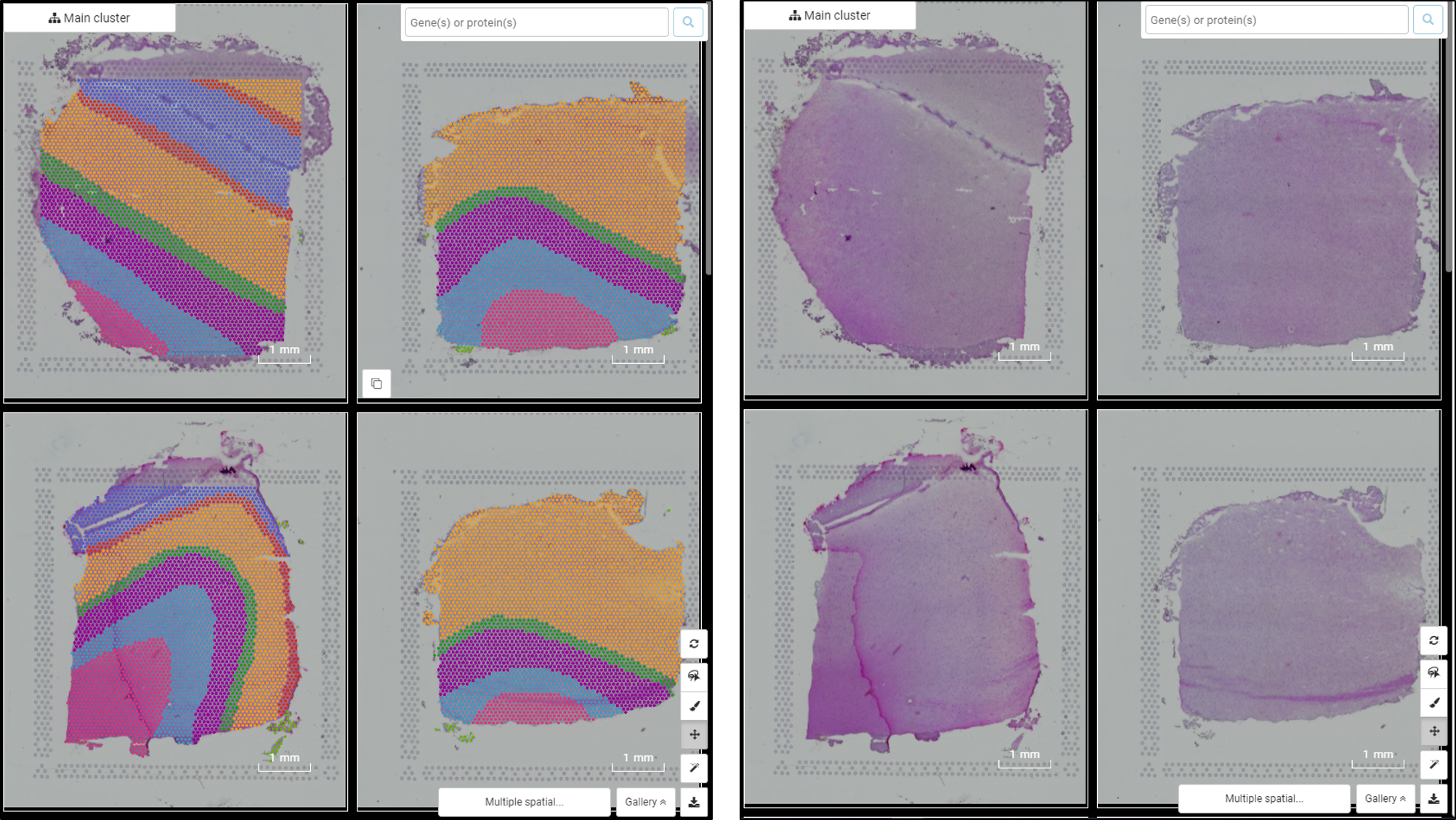
Performing single-cell quality control is a vital step in the data analysis pipeline. Setting QC parameters can significantly affect downstream analysis: too permissive QC filtering makes the dataset too noisy to read, and too stringent QC thresholds may remove important information. In this article, let’s discuss the art of setting […]