Did you know that by the end of 2021, the number of tools for single-cell RNA-sequencing (scRNA-seq) data analysis has passed 1,000? (Zappia and Theis, 2021) This massive resource accelerates the exploration of single-cell data. At the same time, the plethora of options poses several challenges for researchers, such as:
- Too many methods, yet challenging set-up: besides the rapid development of new methods, existing tools also get updated frequently. It’s impossible for users to catch up with the latest scRNA-seq analysis tools. For each method, the setting up of a coding environment is that hardest step. Besides programming skills, time and computer resources are major challenges, to name a few.
- scRNA-seq data is vastly raw and uncurated: the quality of input data greatly influences the final conclusion. While there have been some public databases for scRNA-seq studies, the majority of data remain raw and/or uncurated.
So, how can we overcome these challenges?
BioTuring introduces a solution: The BioTuring Data Science Platform (available at https://datascience.bioturing.com/app/library).
The core idea is simple: we provide you with a platform to run the latest methods for single-cell analysis in R and Python, under the form of ready-to-run Jupyter notebooks, where you can:
- Run the methods without worrying about the environment setup
- Follow our step-by-step tutorials. Just replace them with your parameters and run!
- Access BioTuring massive, curated single-cell database through the BioTuringSDK package. The data has been preprocessed into Scanpy or Seurat objects.
Here are several ways that the BioTuring Data Science Platform accelerates your workflow for single-cell analysis in R and Python.
1. BioTuring Notebook Repository is An Up-to-Date Collection of Single-cell Analysis Methods In R and Python
Do you want to try out novel tools in single-cell analysis, or simply want to catch up with the most recent methods? The platform’s repository is where you can access latest scRNA-seq analysis methods, such as RNA velocity, AUCell, Tangram, CellphoneDB, etc (at the time of writing) in the form of Jupyter notebooks.
We constantly update our collections and are open to users’ requests. Feel free to contact support@bioturing.com to give us suggestions. View the available collection of notebooks at https://datascience.bioturing.com/app/library
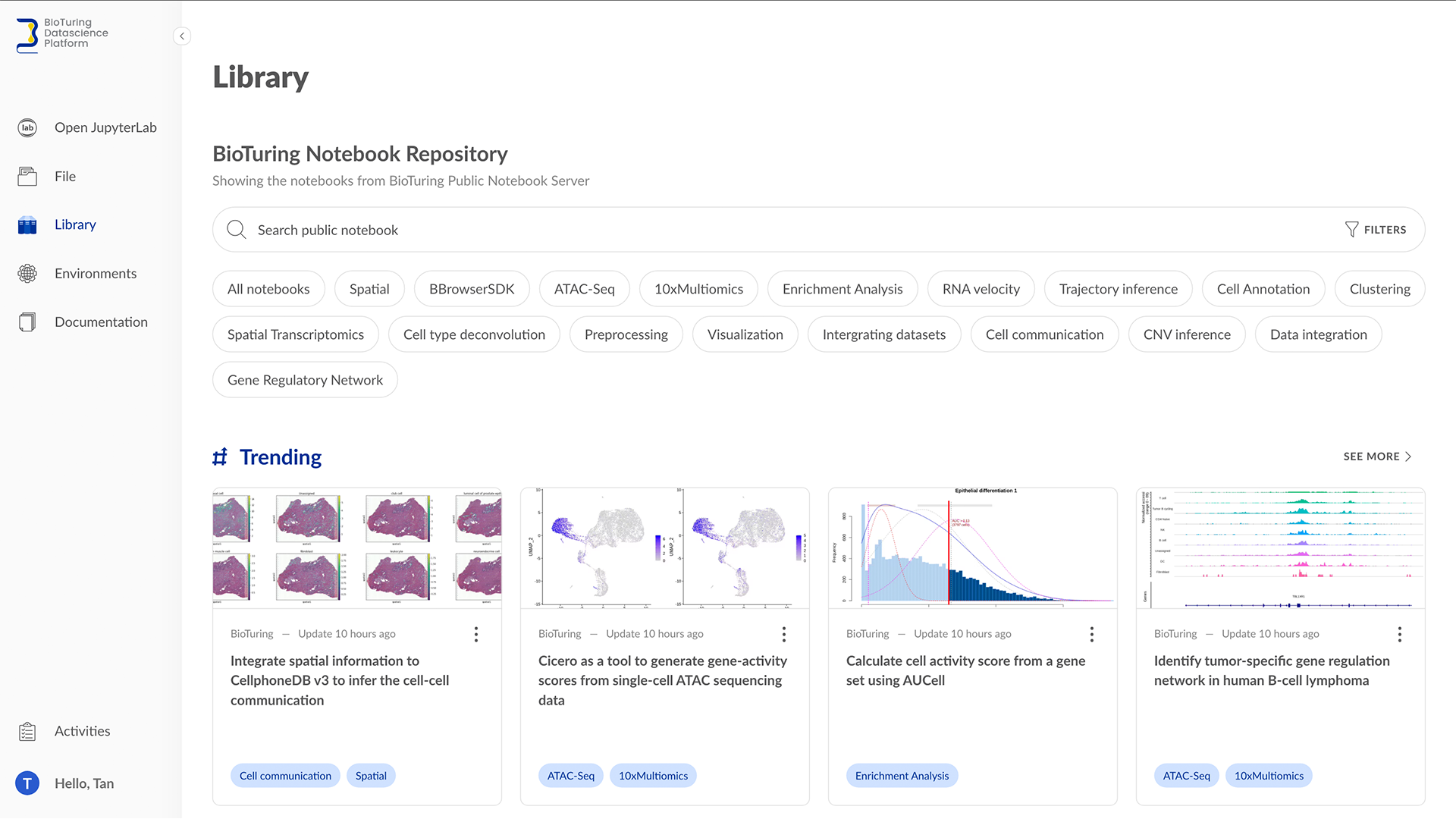
The BioTuring Notebook Repository contains latest scRNA-seq analysis in R and Python
2. Each Single-cell Analysis is a Ready-to-Run Notebook with step-by-step tutorials. No environment setup required
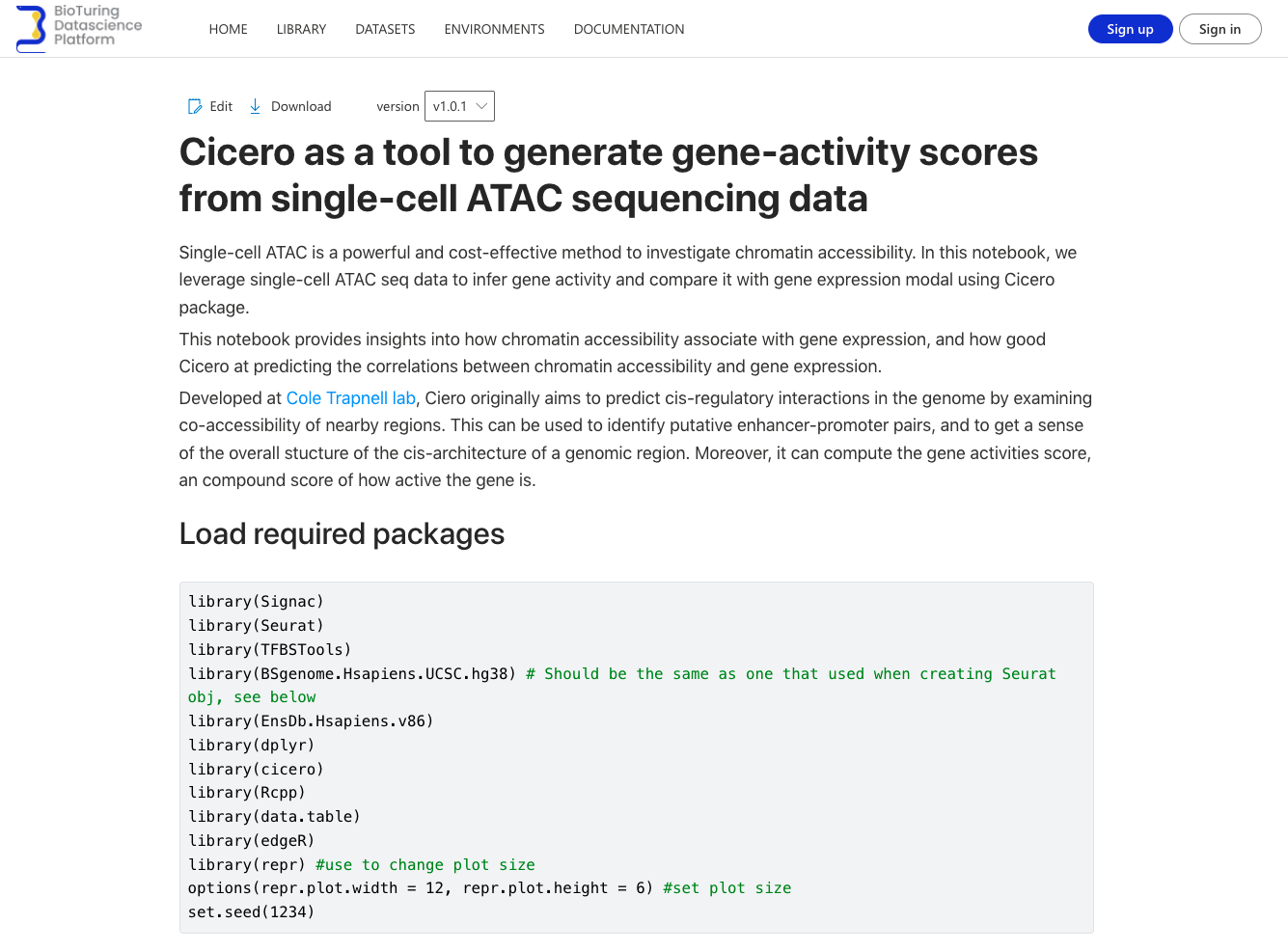
When you run your single-cell analysis in R or Python on the BioTuring Data Science Platform, no environment set up is required. The task has been done by our team. Here’s an example on how to Integrate spatial information to CellphoneDB v3 to infer the cell-cell communication. Users only need to follow our instructions and replace your data when necessary.
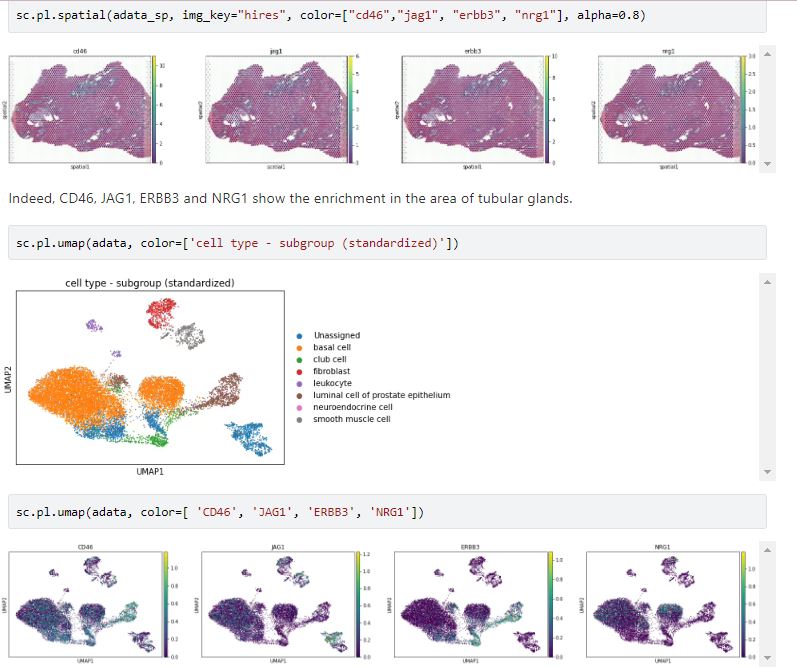
An example: Verify Cellphone DB v3 analysis via the expression of two ligand-receptor pairs ERBB3-NRG1 and CD46-JAG1. The data for cell types, obtained by scRNA-seq, was combined with spatial data by Tangram. Then Cellphone DB v3 searched for ligand-receptor pairs among cells. The whole workflow was done on BioTuring Data Science Platform
You can also create a new environment or import a YAML file – the platform will get the environment ready for you.
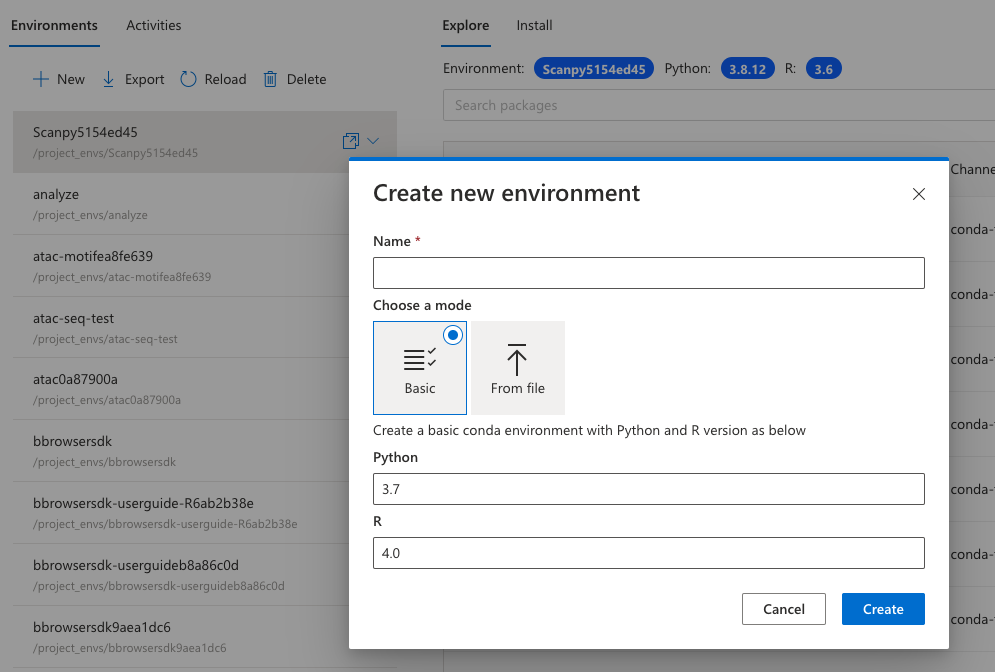
Save your time setting up the environment and go straight to analysis
Each notebook comes with a detailed walk-through, in which we explain how we set up the environment, how to run each step of an analysis (plus examples from real datasets), and how you can modify a certain step with your parameters.
Get guidance for single-cell analysis in R and Python
3. Access BioTuring Curated Single-cell Database
The next advantage of the BioTuring Data Science Platform is the access to ready-to-run input data from the BioTuring Single-cell Database. Containing millions of cells from thousands of studies, the database is comprehensively curated and preprocessed as Scanpy or Seurat objects.They can be loaded to your notebook via the BioTuring SDK R package.
Get curated datasets from the BioTuring Public Database for Single-cell Analysis
4. Increase Reproducibility and Shareability
Finally, the BioTuring Data Science Platform boosts research collaboration by allowing users to share their notebook to others with the environment encapsulated. Recipients can then install the exact same environment and run the shared notebook on our Platform
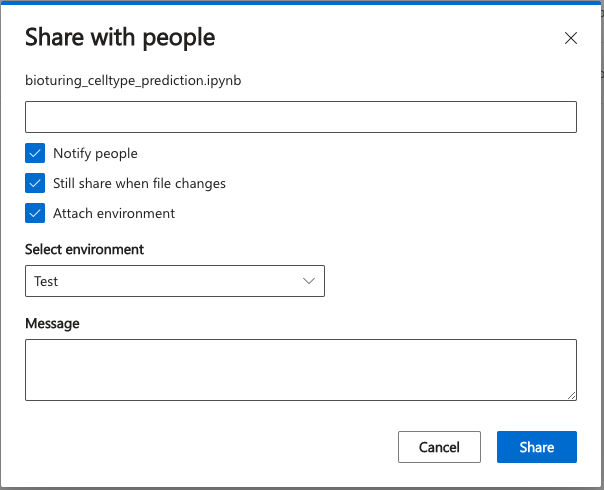
Share your notebook and allow your collaborators to run the same analysis
We hope our solutions meet your needs. The BioTuring Data Science Platform is now available for a trial at https://datascience.bioturing.com/app/library. Feel free to explore the space and let us know what you think!