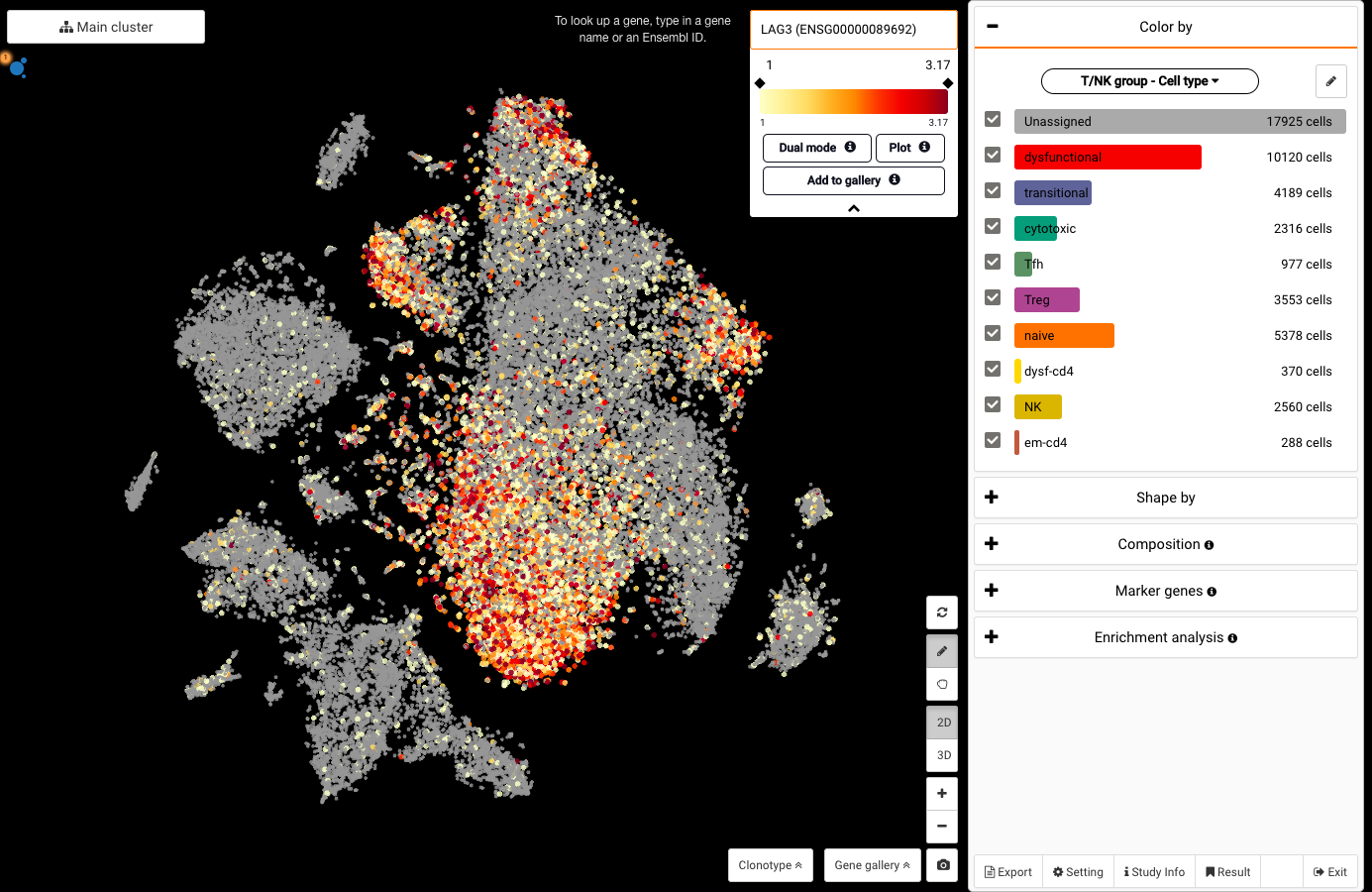
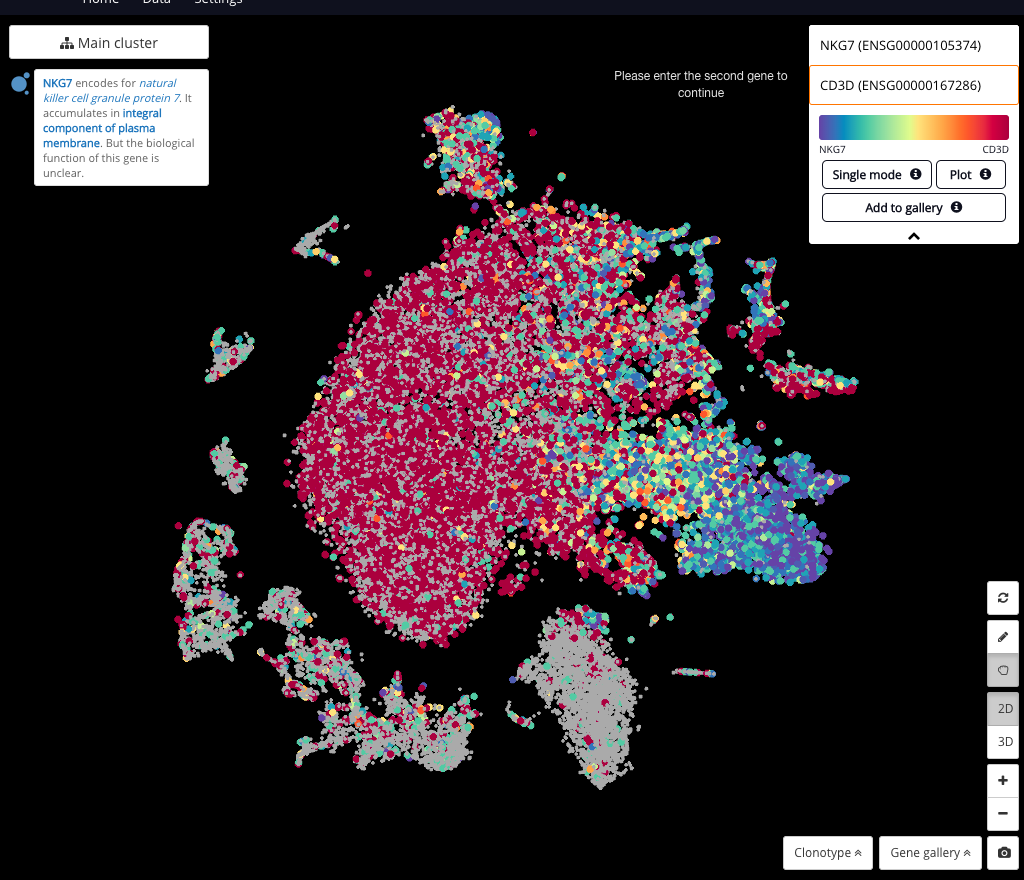
BioTuring Data Science Platform: A Jupyter notebook library of latest methods for single-cell analysis in R and Python
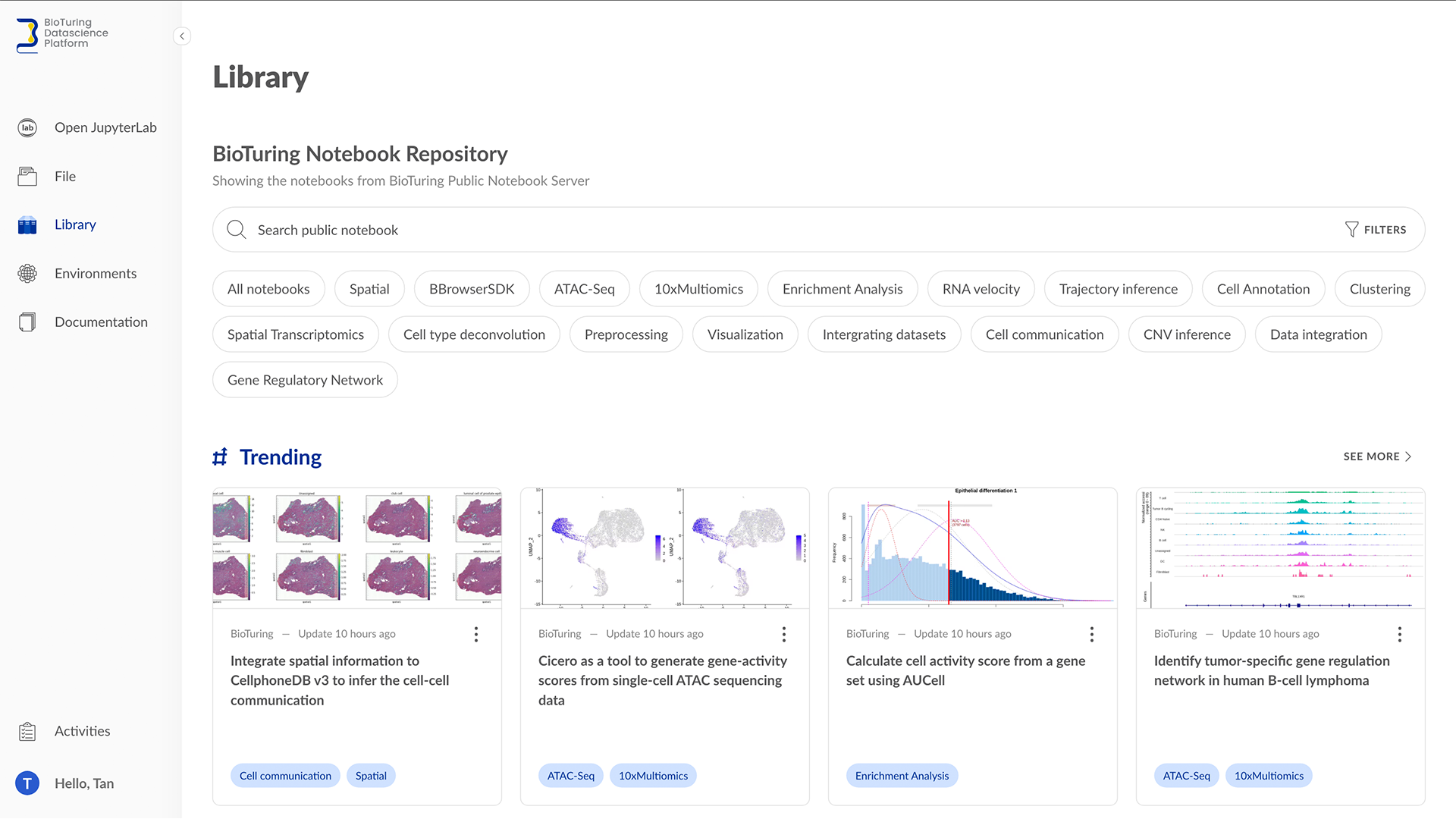
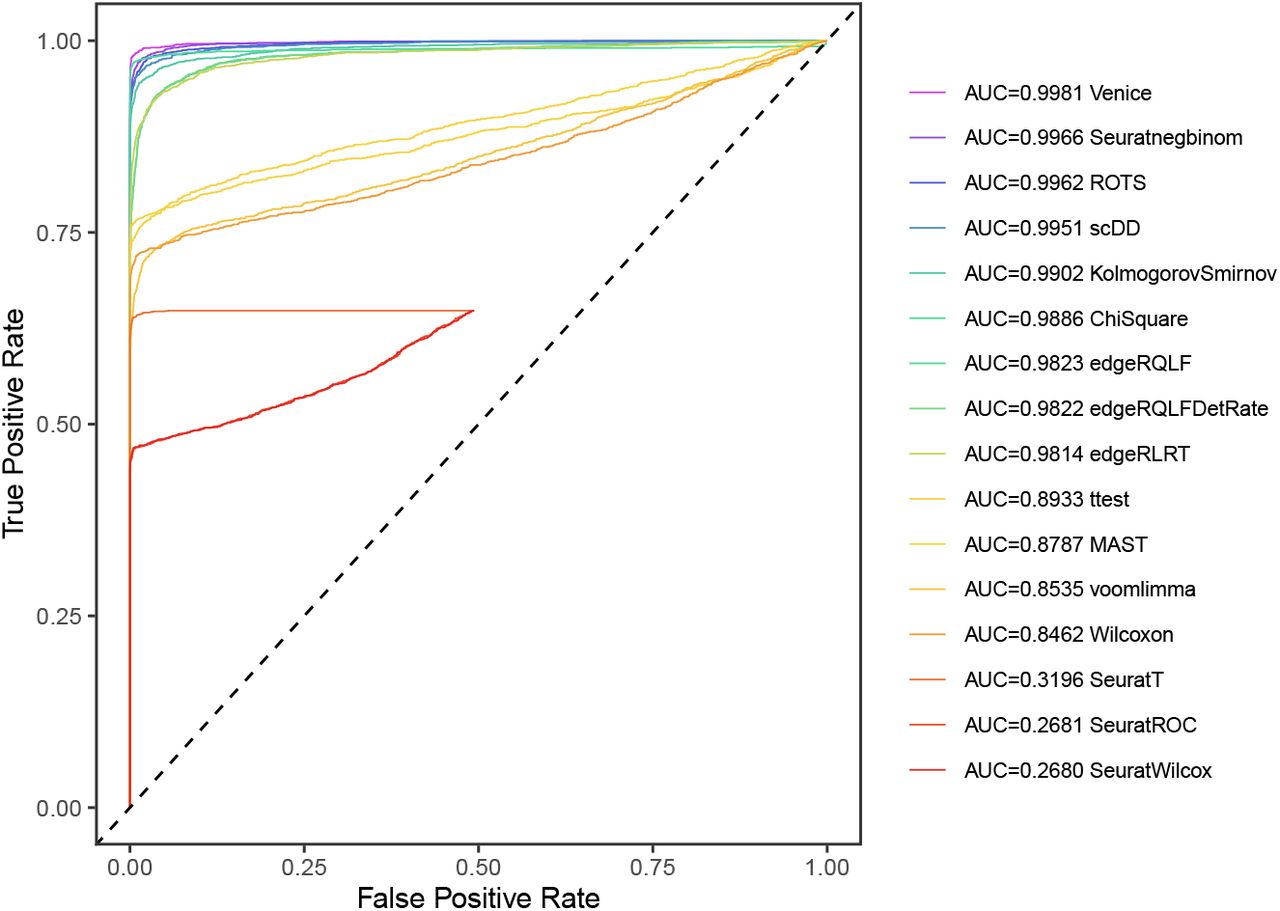
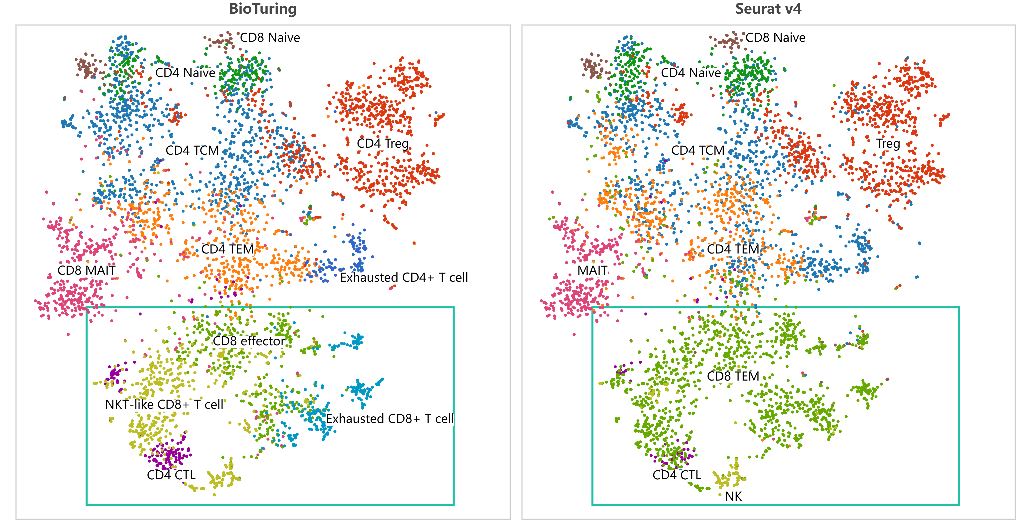
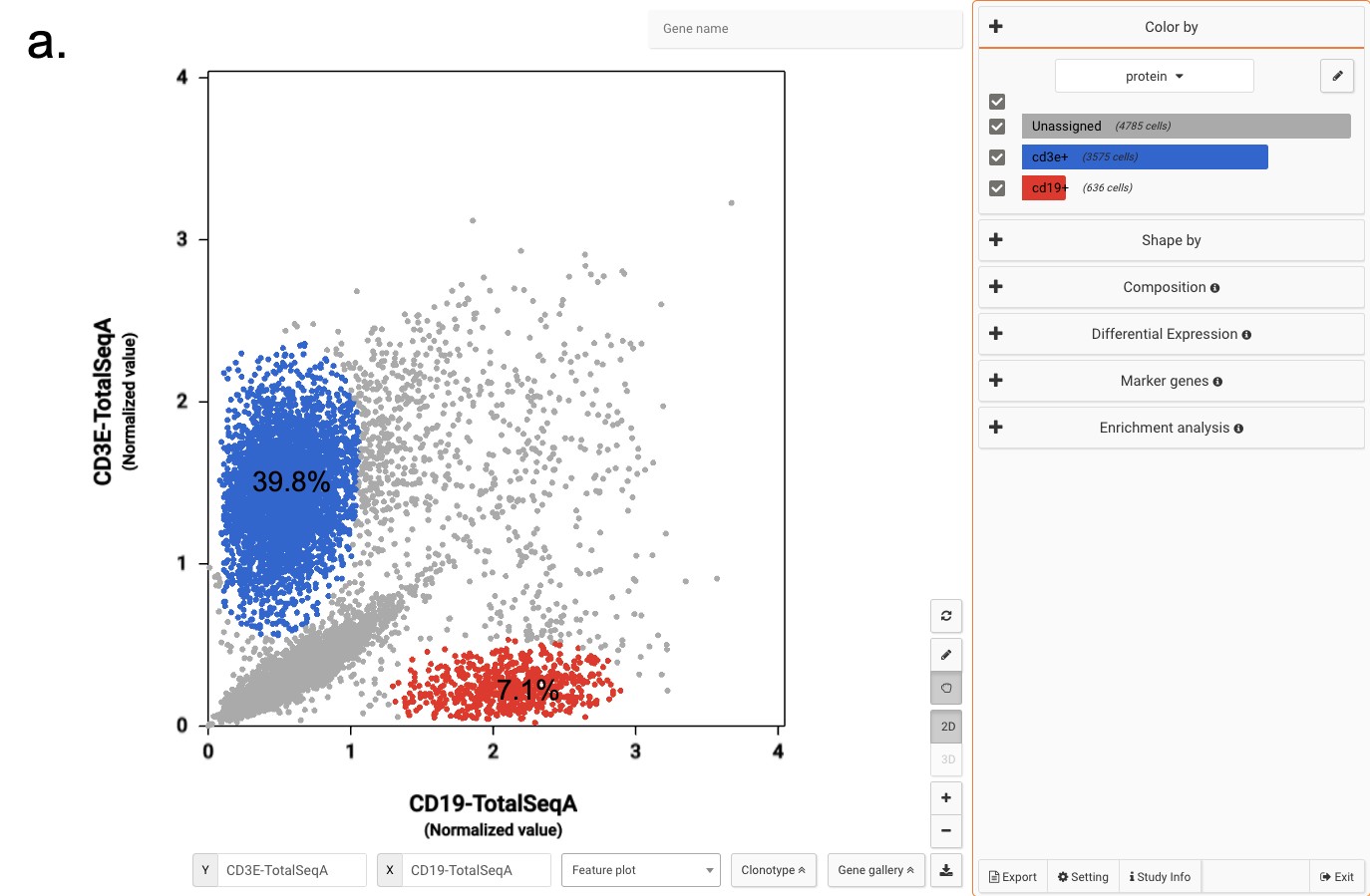
Did you know that by the end of 2021, the number of tools for single-cell RNA-sequencing (scRNA-seq) data analysis has passed 1,000? (Zappia and Theis, 2021) This massive resource accelerates the exploration of single-cell data. At the same time, the plethora of options poses several challenges for researchers, such as: […]