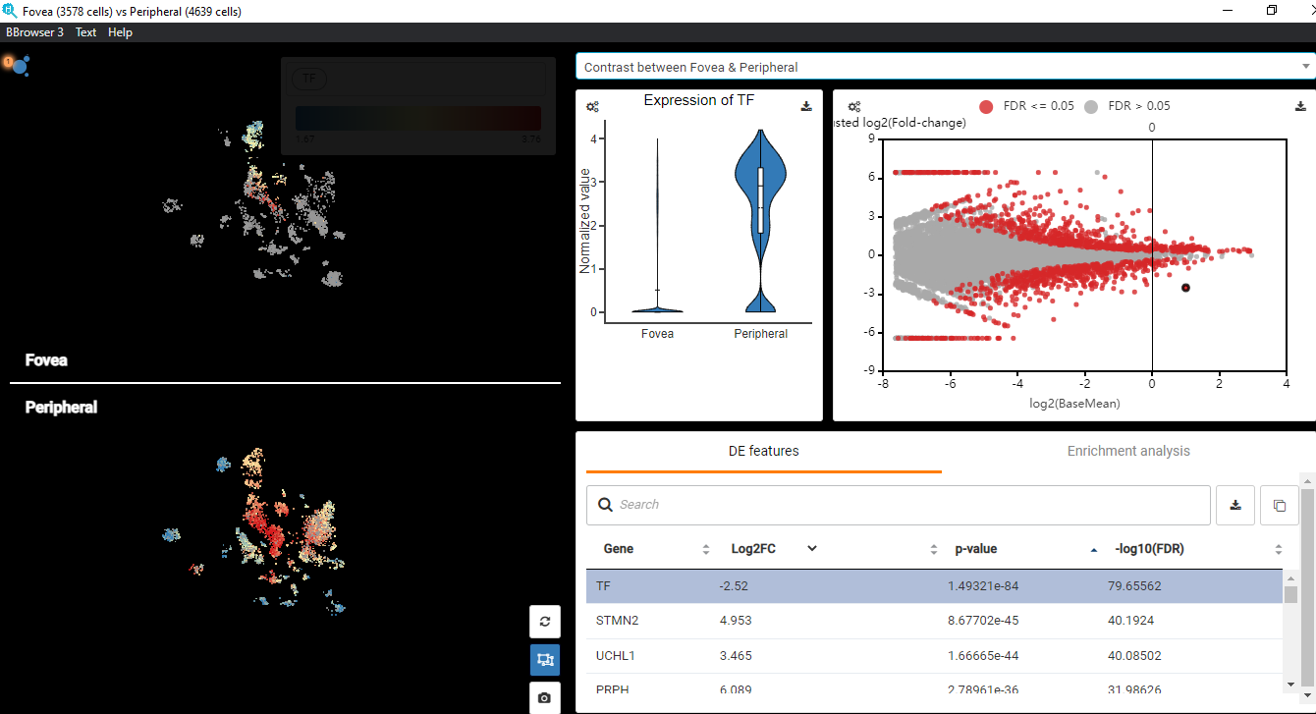
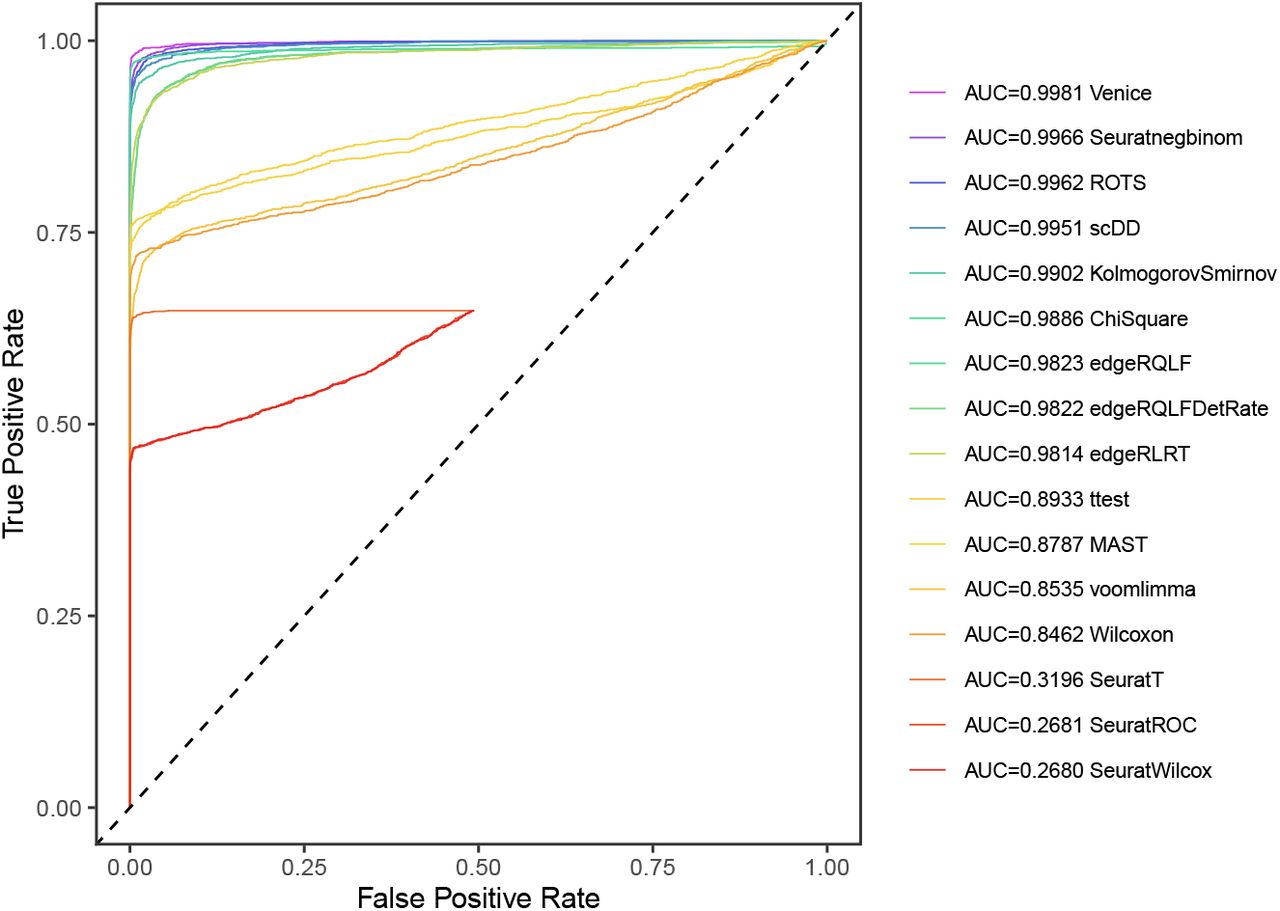
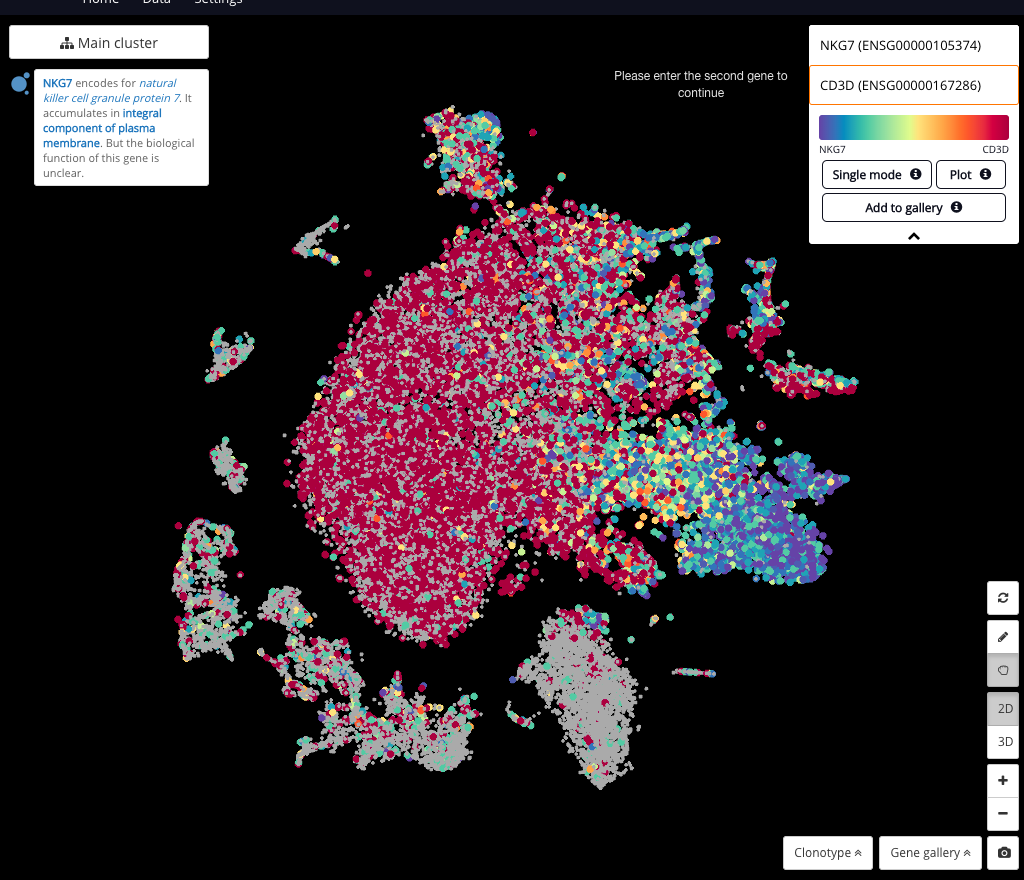
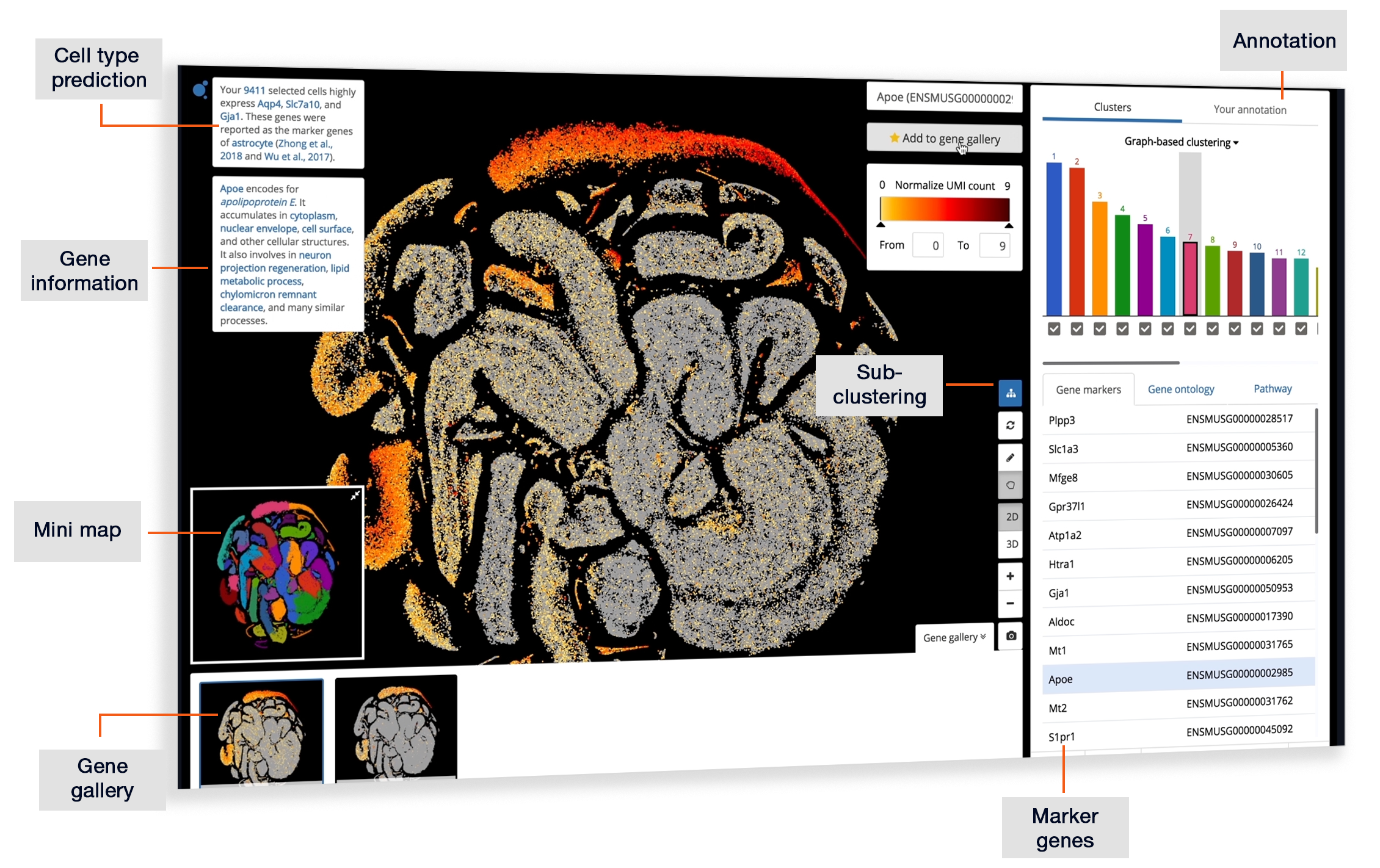
Single-cell RNA-Seq Trajectory Analysis Review
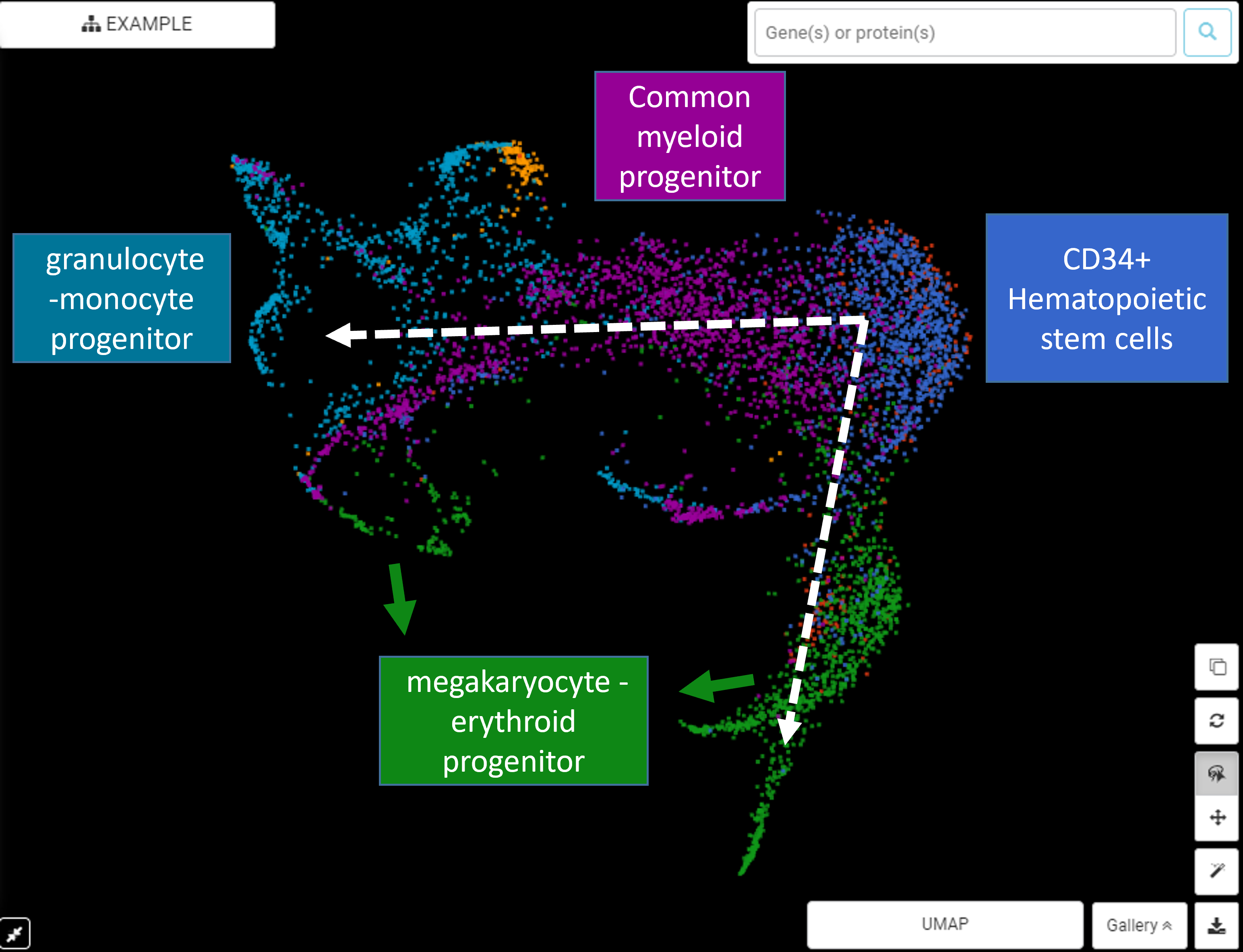
Cellular state often exists in a continuum rather than distinct phases. A prime example is cell development and differentiation. With single-cell RNA-Seq, researchers can observe this continuum of transcriptomic changes via analysis of individual cells. The need to computationally model these dynamics leads to the birth of single-cell RNA-seq trajectory […]