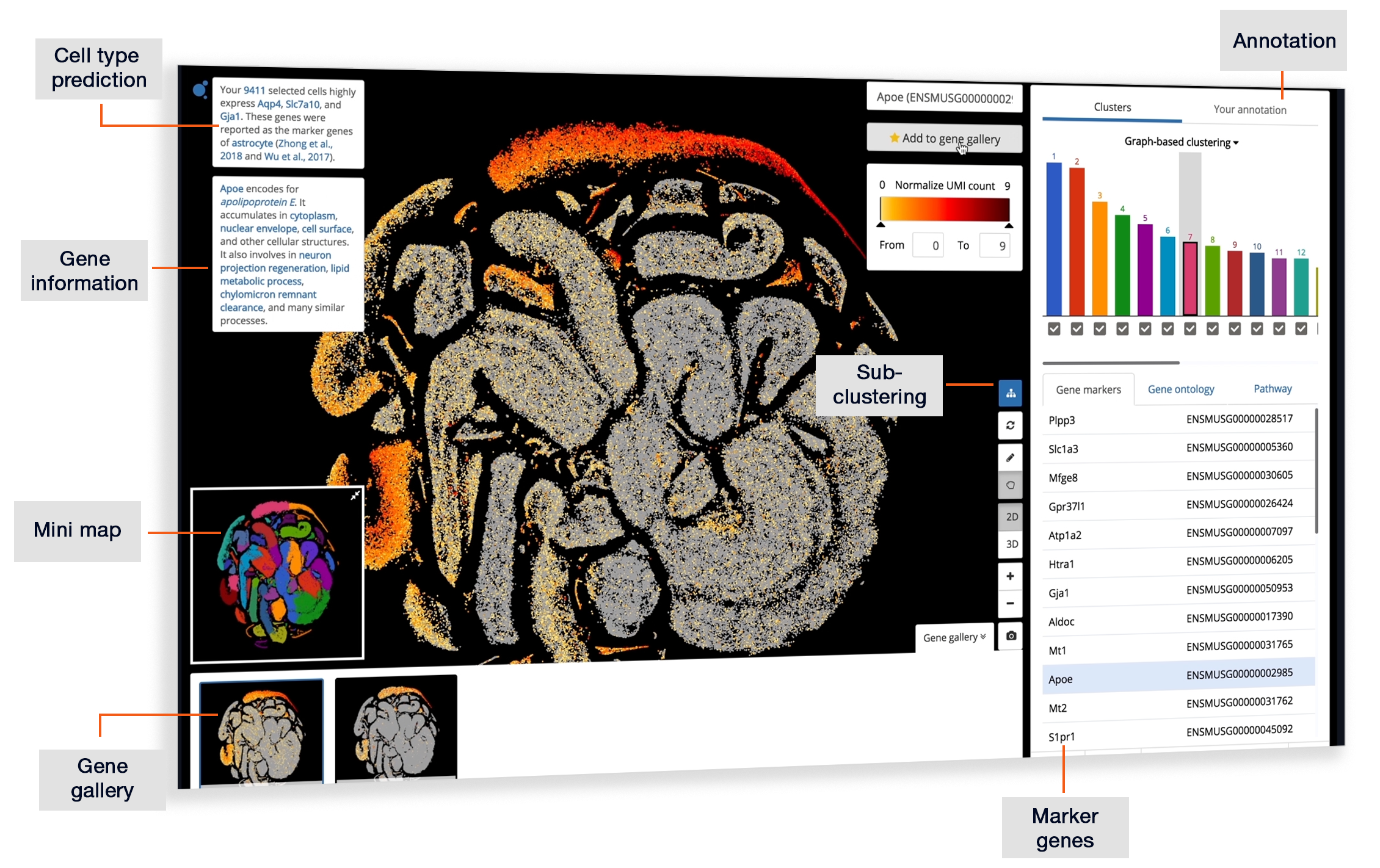
Exploring “Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma” (Li et al., 2018) | BioTuring Cellpedia
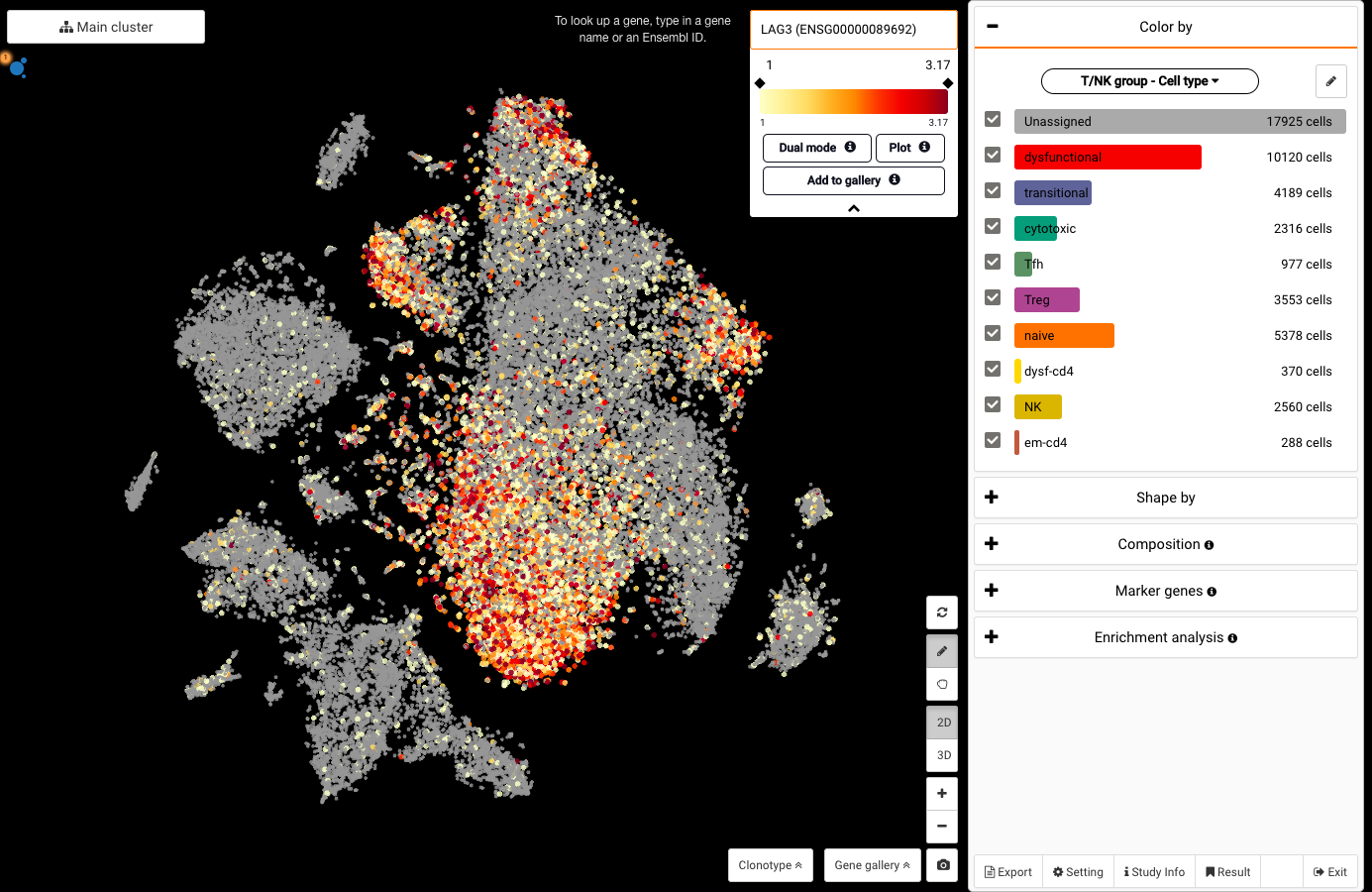
Welcome to our new BioTuring Cellpedia series! We are excited to introduce a new blog series that provides you with an overview of some interesting datasets indexed in BioTuring Browser public repository, a platform for instant access and reanalysis of published single-cell RNA-seq data. Details on their experimental designs and […]